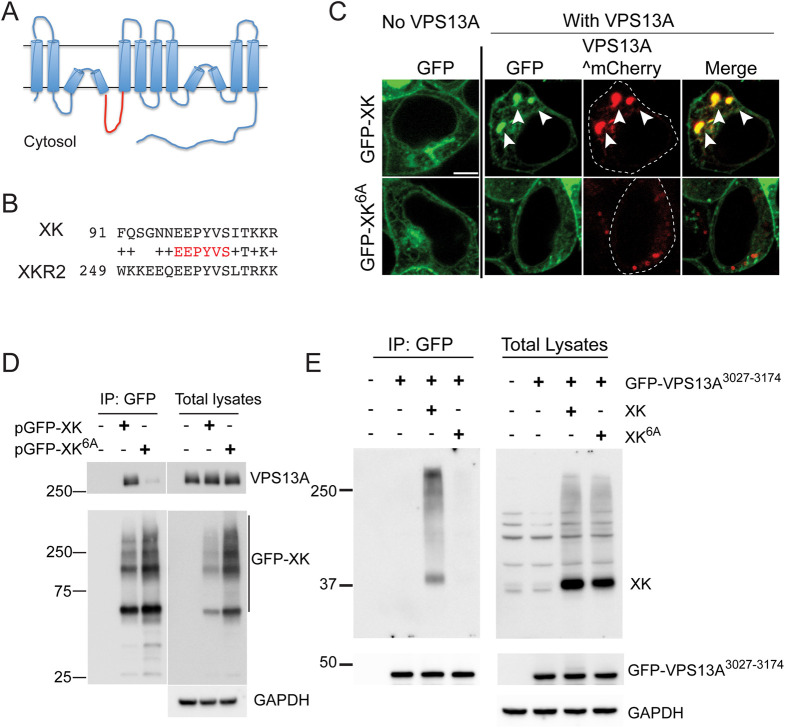
Fig. 4.
Amino acids in the second intracellular loop of XK are required for interaction with VPS13A. (A) Predicted topology of the XK protein based on Alphafold structural prediction (Jumper et al., 2021; Varadi et al., 2022). The second intracellular loop is highlighted in red. (B) Alignment of sequences from the second intracellular loops of XK and XKR2. The invariant residues highlighted in red were mutated to alanines in the XK6A allele. (C) Localization. HEK293T cells were transfected with constructs expressing either GFP–XK or GFP–XK6A with or without VPS13A^mCherry. Arrowheads highlight co-localization of GFP–XK and VPS13A^mCherry. Scale bar: 5 µm. (D) Co-immunoprecipitation of endogenous VPS13A with GFP–XK or GFP–XK6A. HEK293T cells were transfected with constructs expressing either GFP–XK [pcDNA3.1(+)-N-eGFP-XK] or GFP–XK6A (pJS165). GFP–XK was immunoprecipitated with GFP nanobodies as in Fig. 2C. Precipitates and total lysates were probed on immunoblots using anti-VPS13A and anti-GFP antibodies (left panels). GAPDH was used as a loading control. (E) Co-immunoprecipitation of overexpressed untagged XK or XK6A with the VPS13A PH domain. HEK293T cells were transfected with a construct expressing the GFP–VPS13A3027–3174 fusion protein alone (pJS166) or with untagged XK (pJS169) or XK6A (pJS173) and processed as described in Fig. 3A. The experiments in D and E were each performed twice.