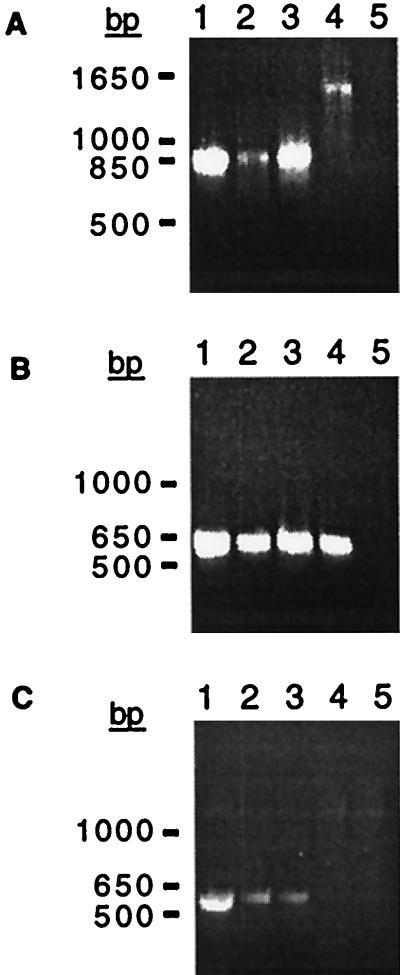
FIG. 5.
PCR amplification of prophage-prophage and chromosome-prophage junctions in classical V. cholerae. (A) Amplication of the junction between a CTX prophage and an upstream element, using primers RstBF2 and RstRproR (Fig. 3). An ∼875-bp product was generated from DNA of classical strains C34 (lane 1), CA401 (lane 2), and O395 (lane 3), indicating that all contain truncated, fused prophages. An ∼1,450-bp product was amplified from the RS1-CTX prophage junction in the O139 strain AS207 (lane 4), and no product was synthesized in the absence of template DNA (lane 5). (B) Detection of a junction between the chromosomal sequence of TLC and a CTX prophage, using primers TLCF1 and Ig1R1. A ∼600-bp product was amplified from the DNA of classical strains C34 (lane 1), CA401 (lane 2), and O395 (lane 3) as well as the O139 strain AS207 (lane 4); it was not synthesized if template DNA was omitted (lane 5). (C) Detection of a junction between CTX prophages in the classical strain-specific insertion site and upstream chromosomal sequence, using the primers PyrFF1 and Ig1R1. A ∼550-bp product was amplified from DNA of the classical strains O395 (lane 1), C34 (lane 2), and CA401 (lane 3); no product was amplified from AS207 DNA (lane 4), which lacks a CTX prophage at this insertion site, or in the absence of template DNA (lane 5).