FIG. 6.
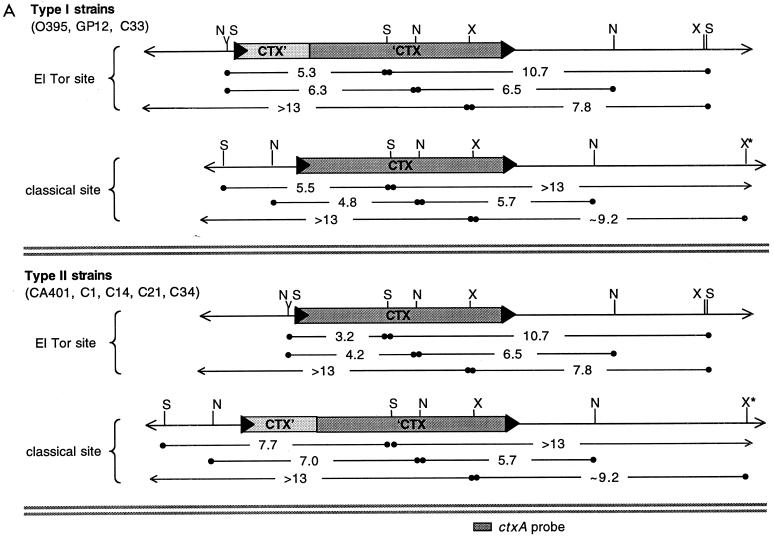
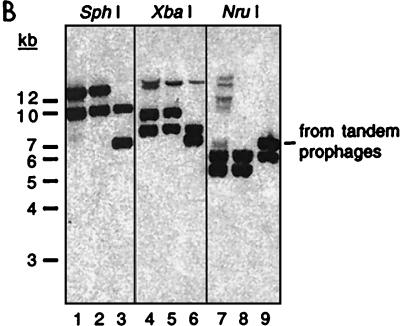
Restriction mapping of the CTX prophage insertions sites in classical biotype strains. (A) Summary of the restriction fragments detected on Southern blots of DNA from type I and type II classical strains digested with SphI (S), NruI (N), and XbaI (X) and probed with various prophage-derived probes. The precise sizes of restriction fragments (in kilobases) were derived from the V. cholerae genome database as well as laboratory sequence data. Band sizes on blots could not be determined as precisely, but all except one were consistent with sequence-derived sizes. X★ denotes an XbaI site that is farther downstream than expected; classical biotypes apparently lack an XbaI site that is present in this region in El Tor strains. The location of the ctxA probe used for panel B is shown at the bottom. (B) ctxAB-spanning restriction fragments from diverse classical strains of V. cholerae are identical. A Southern blot of DNAs digested with SphI, XbaI, and NruI, as indicated, was probed with a ctxA probe. Each enzyme produced equal-size fragments from the classical strains C21 (lanes 1, 4, and 7) and O395 (lanes 2, 5, and 8). AS207 DNA (lanes 3, 6, and 9) had one fragment in common with the classical DNAs for each enzyme. Digests of classical DNA do not have the band with a constant size (∼7 kb), indicative of tandem prophages, that is produced from AS207 DNA. The AS207 prophage array was previously shown to contain an RS1 followed by three tandem CTX prophages. Faint bands in a few lanes resulted from incomplete digestion of DNA.