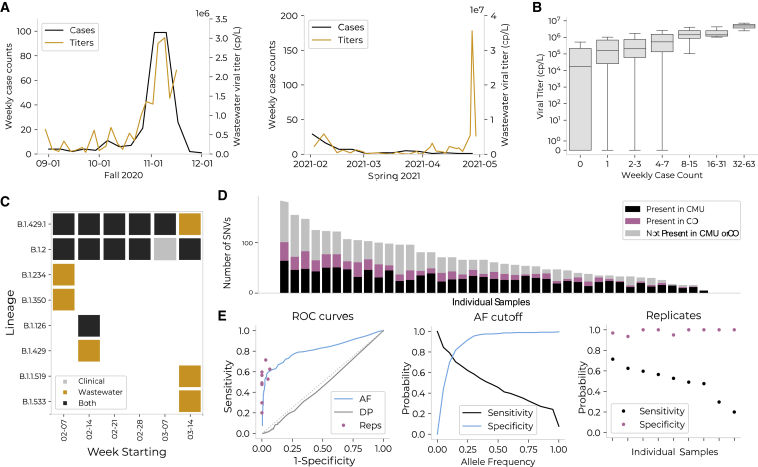
Figure 5.
Wastewater surveillance and sequencing measures aggregate viral load, identifies circulating lineages, and parallels viral genomes from contemporaneous clinical cases
(A) Average wastewater viral titers (orange) versus weekly residential case count (black). Residential case counts were calculated relative to the subsets of dorms monitored (75% of residential population in Fall 2020; 99% in Spring 2021). There was an anomalous peak in wastewater viral titer observed in April, which may be due to technical error, differential shedding patterns, or undiscovered positive individuals.
(B) Viral titer (y axis) versus binned weekly case count (x axis; binned by powers of 2) for each wastewater sample. Viral titer and case count were significantly associated via Fisher’s exact test (on binned slopes; p = 0.04) and Spearman’s correlation = 0.40 (p < 0.001).
(C) Lineages detected on campus via wastewater or clinical sequencing.
(D) The number of single-nucleotide variants (SNVs) detected in wastewater samples; each bar represents a single sample. Individual samples are organized on the x axis in order of total number of SNVs. For each sample, SNVs are categorized by whether they were present in clinical sequences from CMU (black), in clinical sequences from Colorado (pink), or in neither (gray). On average, 51% of SNVs in a single wastewater sample were not found in CMU clinical samples, and 36% were not found in Colorado clinical samples.
(E) Comparison of quality control methods to remove SNVs not validated by presence in Colorado clinical sequences. The three methods compared are: (1) allele frequency (AF): discarding SNVs present at an allele frequency below a given threshold; (2) read depth (DP): discarding SNVs located at a site with a read depth below a given threshold; and (3) replicates (Reps): discarding SNVs not present within both of two technical replicates of a given sample. These analyses are limited to the nine samples for which technical replicates exist. Left: ROC curves for each of the three filters. Middle: sensitivity and specificity for allele frequency-based quality control method. Right: sensitivity and specificity for replicate-based quality control method.