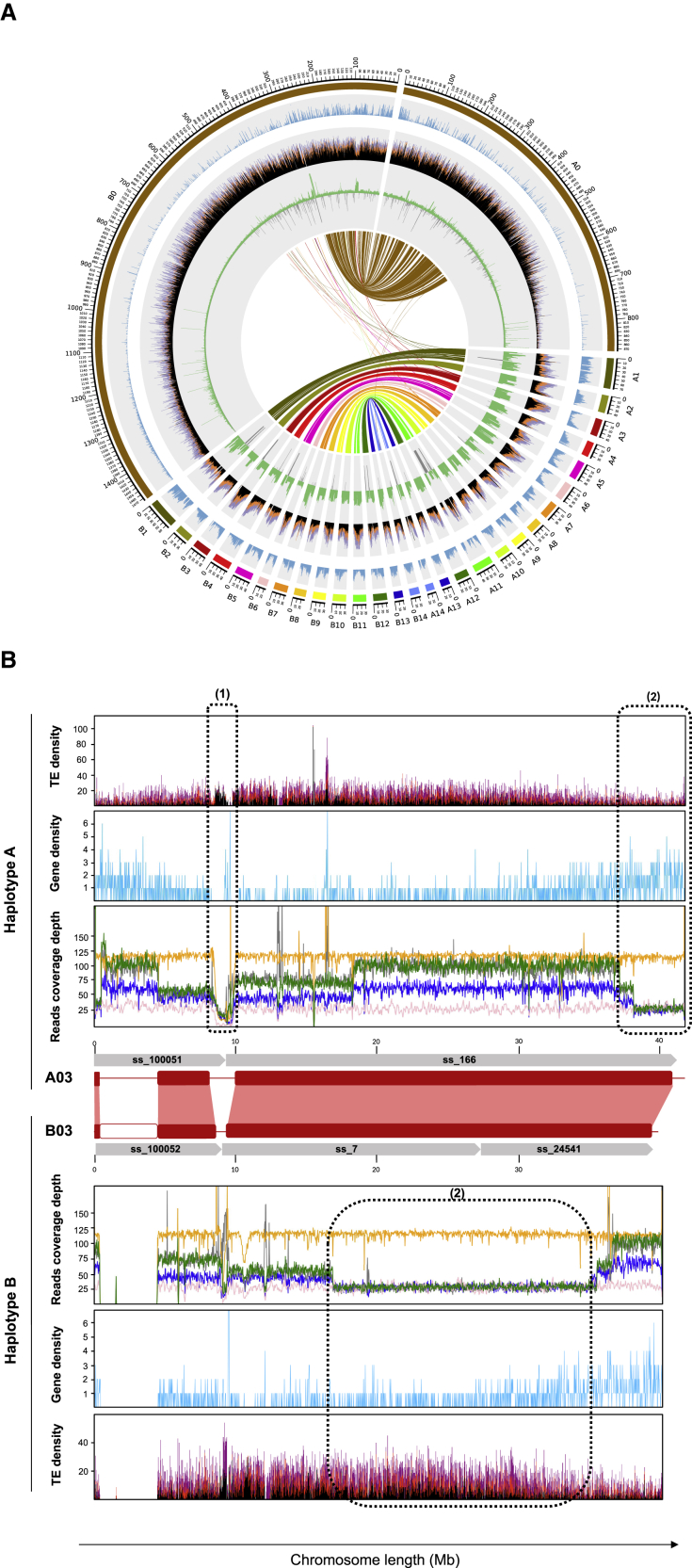
Figure 4.
Overview of the assembled vanilla genome.
(A) Circos plot of the genomic content along V. planifolia haplotypes A and B and the relationship between them. All tracks are divided into 500 kb genomic windows. From the outside to the inside of the circular representation, ideograms of 28 chromosomes and two random mosaic chromosomes that contain the unanchored scaffolds are shown. Gene density (blue) and interspersed repeat RepeatMasker hit density (black: retroelements; orange: long terminal repeat/Copia; purple: long terminal repeat/Gypsy) are shown. Sequencing depth was obtained by mapping CR0040 PacBio HiFi reads on the assembly (green) and N density (gray). Syntenic blocks across haplotypes are connected by lines in the innermost part of the figure.
(B) Sequencing depth along the CR0040 A03 and B03 chromosomes (red rectangles) obtained by mapping Daphna Illumina (yellow) and ONT (pink) reads and CR0040 PacBio HiFi (blue), Nanopore (green), and Illumina (gray) reads onto the CR0040 assembly. Synteny between homologous chromosomes is represented by red boxes. Gaps (N stretches) that explain sudden drops in sequencing depth are shown with white blocks. (1) Low level of sequencing depth for all data is shown. (2) Inverted level of sequencing depth for CR0040 between haplotypes A and B and constant level of sequencing depth for both Daphna haplotypes are shown. Gene and retrotransposon distributions along the chromosomes are represented by a blue line chart and a stacked histogram (Copia: red; Gypsy: purple; other retrotransposons: black), respectively.