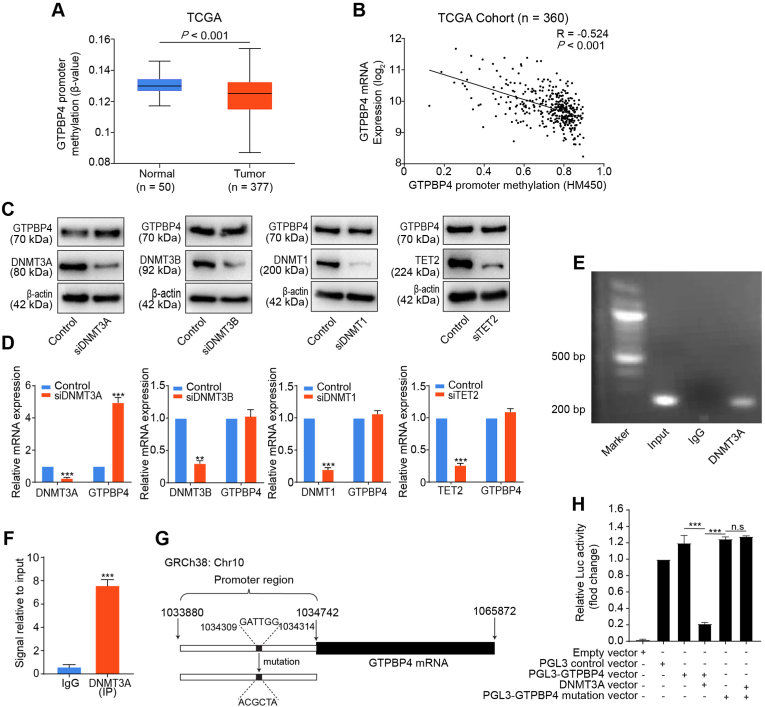
Fig. 1.
Promoter hypomethylation upregulated GTPBP4 expression. (A) GTPBP4 promoter methylation status in normal liver and tumor specimens from TCGA. unpaired 2-tailed t-test. (B) The GTPBP4 promoter methylation status inversely correlates with GTPBP4 mRNA expression in tumor tissues in TCGA. Pearson's correlation test. (C–D) PLC/PRF/5 cells were transfected with DNMT3A, DNMT3B, DNMT1, and TET2 specific siRNAs, respectively. Western blotting (C) and qRT-PCR (D) were used to analyze GTPBP4 expression. n = 3, unpaired 2-tailed t-test. (E–F) ChIP-qPCR analysis of DNMT3A binding to the GTPBP4 promoter region in PLC/PRF/5 cells. The chromatin of PLC/PRF/5 cells was immunoprecipitated with anti-DNMT3A antibody (E), and the GTPBP4 promoter region was amplified by qRT-PCR (F). n = 3, unpaired 2-tailed t-test. (G) The vector containing the predicted binding site in the GTPBP4 promoter control or mutation constructed for luciferase reporter assays. (H) PLC/PRF/5 cells were co-transfected with the pGL3-GTPBP4-WT or mutation reporter and DNMT3A overexpression vector, or its mutation as the control. Dual luciferase activity was determined at 48 h after transfection. Error bars represent the mean ± SD from six independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. n.s, not significant. n = 5, unpaired 2-tailed t-test. Abbreviations: siRNA: small interfering RNA; ChIP-qPCR: Chromatin immunoprecipitation-qPCR.