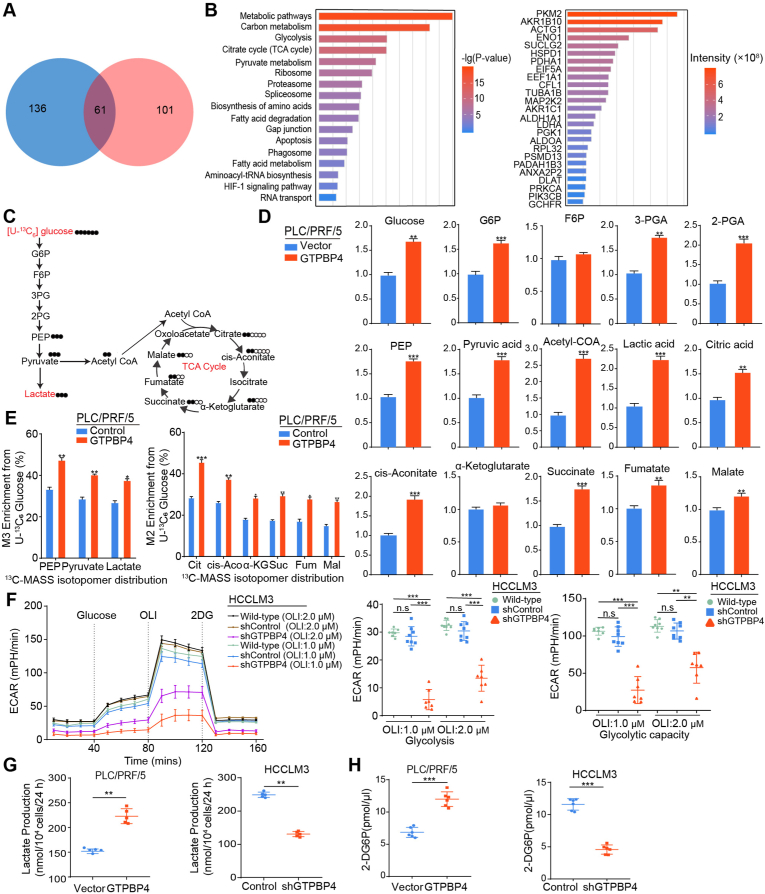
Fig. 3.
GTPBP4 promotes aerobic glycolysis in HCC. (A) Venn diagram illustrating the proteins identified as GTPBP4 binding proteins from GTPBP4 OE PLC/PRF/5 and GTPBP4 OE SMMC-7721 cell groups. (B) Enrichment analysis and functional classification of GTPBP4 specific binding proteins using IPA software (left). The 25 most abundant proteins are listed (right). (C) Flux map and 13C labeled glucose tracing of glycolysis and the TCA cycle. (D) Quantification of intermediate metabolites in glycolysis and the TCA cycle by gas chromatography and mass spectrometry, Q-Exactive-Plus mass-spectrometer, and liquid chromatography coupled with tandem mass spectrometry for different metabolites. n = 6, unpaired 2-tailed t-test. (E) Quantification of isotopes derived from 13C6-labeled glucose intermediates in glycolysis and the TCA cycle was performed using mass spectrometry analysis. M+3 indicates two carbon-labeled PEP, pyruvate, and lactate; M+2 indicates citrate, cis-aconitate, succinate, fumarate, and malate. n = 6, unpaired 2-tailed t-test. (F) The extracellular acidification rate was determined in control and GTPBP4 knockdown HCCLM3 cells. n = 6, one-way ANOVA. (G–H) Lactate production (G) and glucose consumption (H) were measured in HCC cells. Error bars represent the mean ± SD from six independent experiments. n = 6, unpaired 2-tailed t-test. *P < 0.05, **P < 0.01, ***P < 0.001. Abbreviations: IPA: ingenuity pathway analysis; TCA cycle: tricarboxylic acid cycle; ECAR: Extracellular acidification rate.