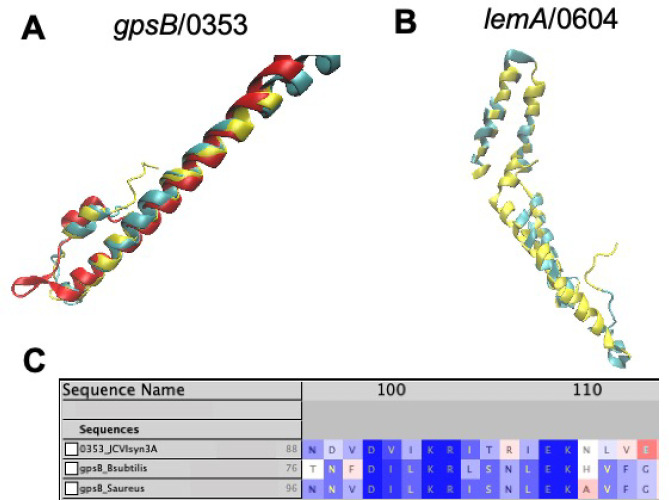
Figure 1.
(A) Predicted structures of gpsB/0353 from AlphaFold2 (red) and RoseTTAFold (blue) aligned with the experimental structure of gpsB from B. subtilis (RCSB PDB: 4UG3)58 at the N-terminal coiled-coil domain using STAMP.41 (B) Predicted structure of lemA/0604 (see section 3) from RoseTTAFold (blue) aligned by FATCAT25 with the lemA two-component cell growth regulator from T. maritima (yellow, RCSD PDB: 2ETD(61)). 142 of the 210 residues (68%) are well-aligned with a RMSD of only 1.67 Å. (C) Multiple sequence alignment of /0353 from JCVI-syn3A with sequences of gpsB from B. subtilis (strain 168) and S. aureus (strain NCTC 8325/PS 47) shows the conserved C-terminal domain consensus sequence,59 lending further evidence to the assignment of gpsB/0353 instead of its paralogue divIVA. All images visualized in VMD.42