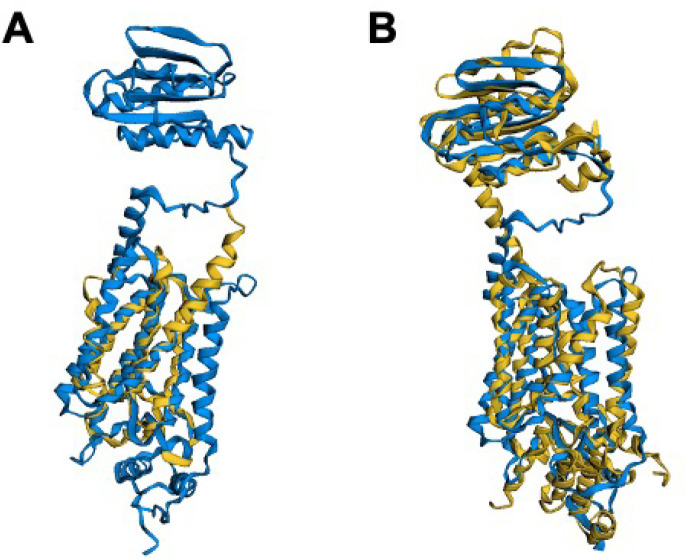
Figure 6.
(A) FATCAT25 structural alignment of AlphaFold2 predicted structure of /0516 (blue) and gplG protease from H. influenzae (RCSB PDB: 2NR9(89)), where only the lower helical portion of the protease is available in the experimental structure. (B) FATCAT structural alignment of the predicted structure from (A) with the Alphafold2 predicted structure for the gplG paralogue protease yqgP from B. subtilis shows excellent agreement to both the upper β- sheet and the lower -a-helical regions of the JCVI-syn3A structure, with a p-value of 7.19 × 10–9 showing significant structural similarity.