Figure 8.
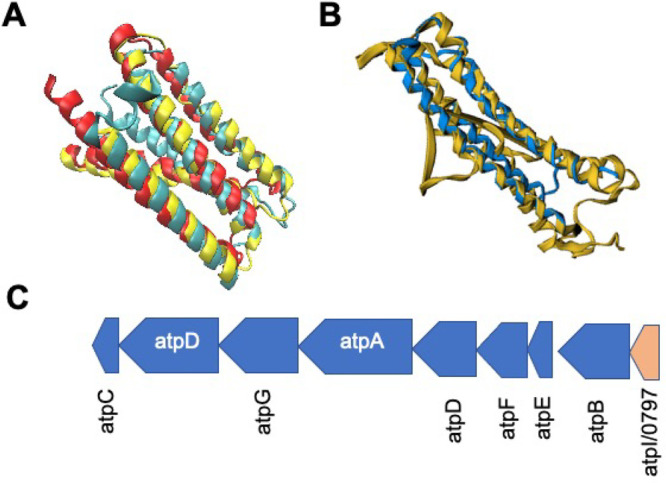
(A) STAMP structural alignment of predicted structure for B. subtilis atpI (yellow) with predicted protein structures for atpI/0797 from AlphaFold2 (red) and RoseTTAFold (blue). Images generated using VMD.42 Alignment of these two structures obtained a FATCAT25p-value of greater than 1.0 × 10–11, showing significant similarity with 92% of the JCVI-syn3A structure being well-aligned to that predicted for B. subtilis. (B) FATCAT alignment of the Spinicia oleracea atpI structure (RSCB PDB: 6FKI(101)) with the AlphaFold2 predicted structure of atpI/0797 with conservation of the central two α-helical regions especially. Alignment of these two structures obtained a FATCAT25 gave 130 residues or 99% of the JCVI-syn3A structure being well-aligned to the larger 247 residue cryo-EM structure for Spinicia oleracea. (C) Genomic organization from JCVI-syn3A and B. subtilis of the ATP synthase operon3,49 is identical lending further support to the assignment of atpI/0797 (red, bottom-right).
