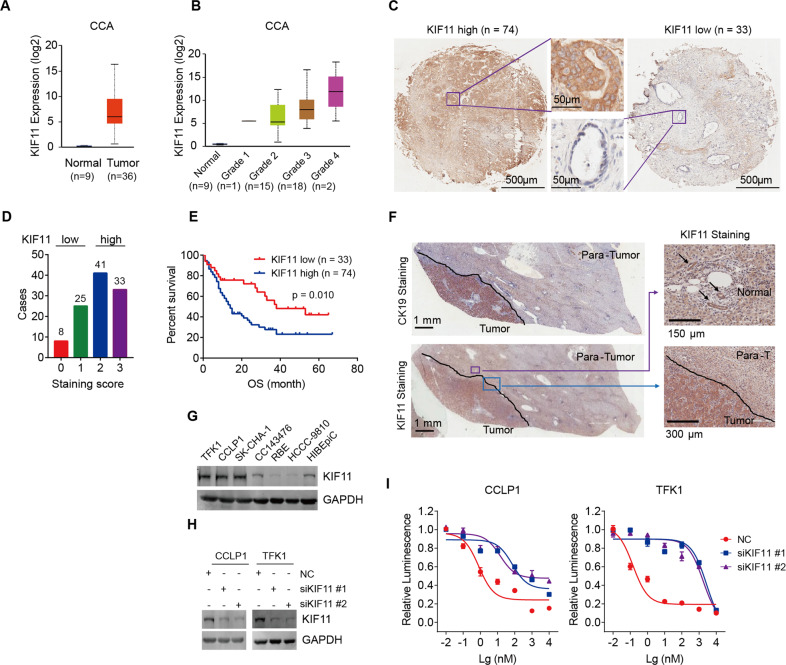
Fig. 3. KIF11 gene expression and dependency in CCA.
A KIF11 expression in CCA patient samples compared to that in normal tissue samples within the TCGA dataset. Statistical analysis was performed using the two-sided unpaired t-test. B KIF11 expression in CCA patient samples grouped based on pathological grade. Statistical analysis was performed using the two-sided unpaired t-test. C Representative images of KIF11 high- and low-expression in human CCA tumor tissue samples by immunohistochemical analysis. D The number of cases in the KIF11 low and high groups. Human CCA samples with staining scores of 0 and 1 were allocated to the KIF11 low-expression group (n = 33), whereas those with a score of 2 and 3 were allocated to the KIF11 high-expression group (n = 74). E Overall survival of CCA patients with high and low KIF11 gene expression using Kaplan-Meier survival curves. Statistical analyses were performed using the log-rank test. F Representative images of KIF11 expression in tumor, para-tumor, and normal tissues from CCA patients by immunohistochemical analysis. Arrows indicate intrahepatic bile ducts. G Expression of KIF11 in CCA cell lines and normal cell line HIBEpiC detected by immunoblotting. H The knockdown efficacy of KIF11 siRNA detected by immunoblotting. I Cell sensitivity to SB743921 treatment in control and KIF11-knockdown cells. CCLP1 and TFK1 cells transfected with indicated siRNA or NC were exposed to SB743921 for 72 h. Cell viability was measured using the CellTiter-Glo luminescent cell viability assay.