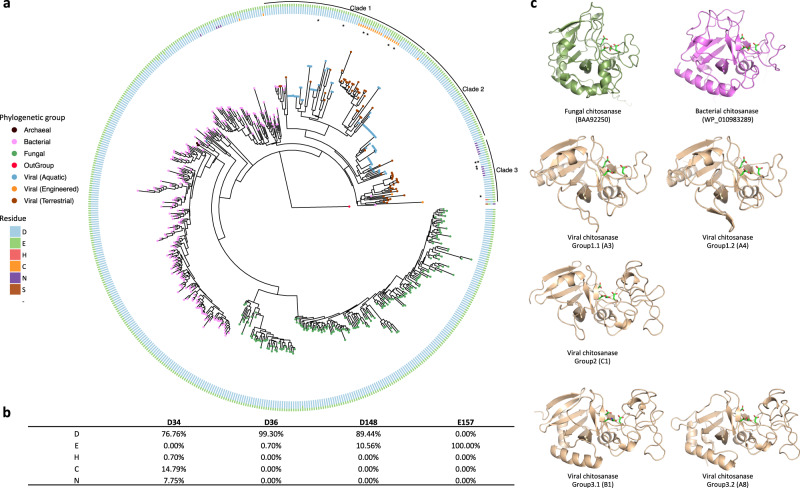
Fig. 1. GH75 chitosanases detected across domains of life.
a The phylogenetic tree with viral, fungal (green), bacterial (pink) and archaeal (black) chitosanases is rooted by a bacteriophage lysozyme (YP_006987285.1, red node). The tree leaves representing viral chitosanases are colored by habitat: aquatic (blue), engineered (orange), terrestrial (brown). The four key residues in the proposed active site are color coded and shown in the circular rings sequentially with the residue closest to the N-terminal position in the innermost ring. The viral chitosanases selected for enzymatic function validation are highlighted with asterisks. The viral chitosanase used for crystallization is labeled with two asterisks. b The relative frequencies of each residue at the four conserved sites were calculated and shown in the table. c The protein structures of representative chitosanases were predicted by AlphaFold20 and are colored according to phylogenetic groups: bacterial (pink), fungal (green), viral (brown).