Heterozygous single nucleotide variants (SNVs) or copy-number variant (CNV) deletions, involving the mesenchymal forkhead box family transcription factor gene, FOXF1, or its distant lung-specific enhancer, are responsible for 80%–90% of cases of alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV).1 ACDMPV is a lethal lung developmental disorder with severe progressive respiratory failure and persistent pulmonary arterial hypertension (Supplemental Material). Intriguingly, in contrast to point mutations in FOXF1, the ACDMPV-causative CNV deletions arise de novo almost exclusively on the maternal chromosome 16q24.1. Thus far, we and others have described 50 de novo CNV deletions that arose on maternal chromosome 16 and only three de novo CNV deletions that arose on paternal chromosome 16q24.1 (Fig. S1). Here, we define an ∼660 bp ultra-conserved non-coding interval (660UCR) within the FOXF1 enhancer as critical for FOXF1 cis-regulation and normal human lung development. Heterozygous loss of this region on paternal chromosome 16 was found causative for ACDMPV. We also describe a novel enhancer lncRNA gene, RP11-805I24.3, located in the proximity to 660UCR and overlapping another ∼1 kb ultra-conserved interval (1000UCR), as essential for the FOXF1 expression. Based on the obtained data, we propose a bimodal structure and parental functional dimorphism of the FOXF1 enhancer.
The lung-specific FOXF1 enhancer in humans has a complex structure, consisting of the proximal Unit 1 and distal Unit 2 (Fig. 1A).1,2 Both units encode regulatory lung-expressed lncRNAs and feature parent-of-origin-specific epigenetic modifications. Using genome sequencing in ACDMPV family trio (204.1–3), we have identified a pathogenic heterozygous, 8.8 kb CNV deletion within Unit 1 that arose de novo on the paternal chr16q24.1 (Fig. 1A, S2–S6). Together with 33 other CNV deletions of the FOXF1 enhancer, this second smallest deletion enabled identification of the ultra-conserved 660UCR interval. Longer stretches of homology with 660UCR can be found in the genomes of lobe-finned fishes, including coelacanth and lungfish, whereas short sequence stretches are visible in some actinopterygian fishes with the vascularized swim bladder (Fig. 1A). For comparison, Unit 2 of the enhancer contains two non-coding intervals which are conserved down to turtles only.
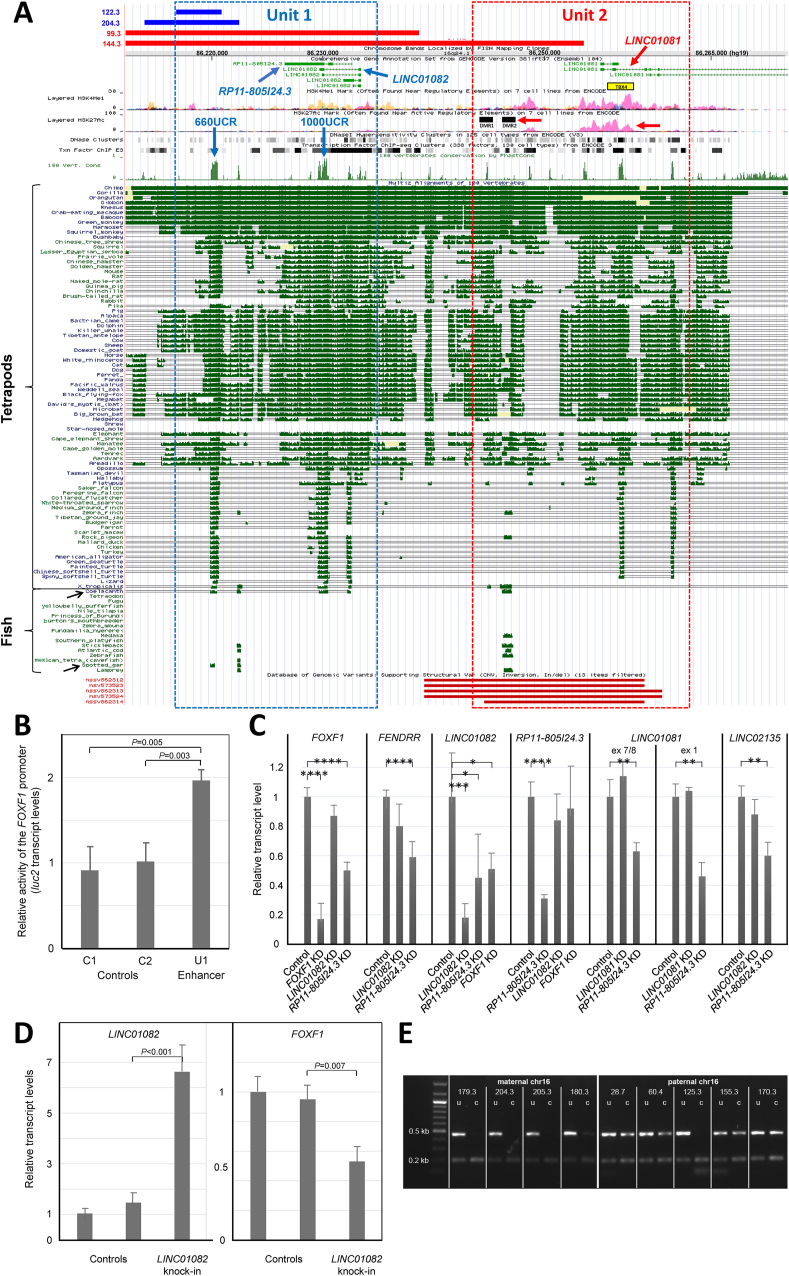
Figure 1.
Bimodal structure and parental dimorphism of the ∼60 kb FOXF1/FENDRR lung-specific enhancer. (A) UCSC genome browser representation of the enhancer, defined by the smallest deletion overlap of 29 pathogenic CNV deletions at chromosome 16q24.1 leaving FOXF1 intact (Fig. S1). On the top are four additional different-sized CNV deletions identified in ACDMPV patients on maternal (red) or paternal (blue) chromosome 16q24.1 that provided further insight into bimodal structure and function of the enhancer. Blue arrows in Unit 1 indicate an ∼660 bp ultra-conserved interval, 660UCR, mapping within the ∼4 kb smallest deletion overlap of 33 CNV deletions, and antiparallel pair of the lung-expressed lncRNA genes LINC01082 and RP11-805I24.3 overlapping another ultra-conserved interval, 1000UCR. The red arrows in Unit 2 indicate (i) the lung-specific lncRNA LINC01081 gene (ii) the interval with the typical epigenetic signatures of an enhancer, and (iii) two differential methylation regions described recently by Slot and coworkers.3 In addition, our ChIP-seq studies showed TBX4 binding to the Unit 2 (yellow rectangle). Black arrows indicate coelacanth and spotted gar fish. At the bottom, there are five benign CNV deletions assessed from DGV, involving Unit 2 of the enhancer, identified in apparently healthy individuals. We suggest these polymorphic deletions may be located on paternal chromosome 16q24.1. (B) Luciferase reporter assay showing the significant increase of FOXF1 promoter activity by juxtaposed fragment of the enhancer Unit 1 containing the ultra-conserved 660UCR interval. (C) Knock-down experiments in IMR-90 cells showing regulatory relationships between enhancer lncRNAs, FENDRR, and FOXF1 (P ≤ 10−n (n = 2∗–4∗) and <0.05 (∗)). (D) Knock-in of LINC01082 showing suppression of FOXF1 by overexpression of this lncRNA. (E) The analysis of the methylation status of the ultra-conserved 1000UCR interval of the FOXF1 enhancer mapping within the overlapping pair of mutually antisense lncRNA genes LINC01082 and RP11-805I24.3. ACDMPV lung (179.3, 28.3, 60.4, 125.3, 155.3, 170.3), blood (204.3, 205.3) and umbilical cord (180.3) tissues were analyzed. PCR was done using undigested (u) and digested (c) DNA.
Applying luc2 reporter assay in fetal lung fibroblasts IMR-90, we have shown that the 660UCR interval increases the activity of the FOXF1 promoter two-fold (Fig. 1B), consistent with the fact that the CNV deletions of the paternal 660UCR allele in pts 204.3 and previously reported 122.3 (Fig. S1) led to full ACDMPV phenotype. A portion (EH38E1835120) of this interval is annotated in ENCODE's Candidate Cis-Regulatory Elements (cCREs) database as having distal enhancer-like signature. In addition, mouse region syntenic to human 660UCR was shown to function as the Foxf1 enhancer.3 The second ultra-conserved region of Unit 1, 1000UCR, is included in GeneHancer regulatory element GH16J086193 and overlaps the lncRNA genes LINC01082 and RP11-805I24.3. Knock-down of these lncRNAs using siRNA in IMR-90 cells, showed that RP11-805I24.3 positively regulates expression of FOXF1, FENDRR, and other analyzed here lncRNAs (Fig. 1C), suggesting that it may function as a general transcriptional activator. On the other hand, LINC01082 (Fig. S7), which has expression of an order of magnitude lower than that of RP11-805I24.3, decreases expression of FOXF1 when up-regulated in IMR-90 cells from the pcDNA-based construct to the level of RP11-805I24.3 (Fig. 1D). Importantly, both 660UCR and 1000UCR bind EP300, a histone acetyltransferase that catalyzes H3K27ac deposition typical for active enhancers and partially overlap the RNA PolII binding site (ENCODE's ChIP-seq database). Moreover, EP300 is known to directly interact with HIF1A in regulation of hypoxia-induced VEGF and with RNA PolII (GeneCards). Using luc2 reporter assay, we have also found that the 660UCR interval does not regulate RP11-805I24.3 promoter (Fig. S8). Thus, 660UCR and RP11-805I24.3 seem to positively regulate FOXF1 expression independently of each other. Applying siRNA knock-down to FOXF1, we sought whether FOXF1 controls expression of its enhancer lncRNAs in a manner similar to how it regulates FENDRR. We have found that FOXF1 positively regulated LINC01082 but not RP11-805I24.3 (Fig. 1C). Thus, there is no evidence for a regulatory feedback loop interaction between FOXF1 and lncRNAs encoded in its enhancer.
Interestingly, Unit 1 of the FOXF1 enhancer was previously shown to be differentially methylated at CpG dinucleotides.2,4 Here, using methylation sensitive digestion assay, we have found methylation of paternal but not maternal allele of 1000UCR (Fig. 1E), whereas both parental alleles of the 660UCR were methylated, although showing a trend towards stronger methylation of the paternal allele (Fig. S9).
Unit 2 features lung-specific epigenetic histone 3 modifications typical of an active enhancer, and includes the 3′ portion of the lung-expressed lncRNA gene LINC01081 (Fig. 1A). In this ∼5 kb-large interval, we have described previously four rare non-coding SNVs mapping in trans to heterozygous pathogenic SNV and CNV deletions involving FOXF1 and/or its enhancer, that, likely acting as hypermorphs, might have ameliorated the lethal ACDMPV phenotype. Interestingly, Unit 2 was also reported to exhibit features of differential CpG methylation (SNP rs1621902)2 and partially overlaps ChIP-seq-determined TBX4 binding region (Fig. 1A). Moreover, it features putative allelic differences of the H3K27Ac profile and the presence of two differentially methylated regions (Fig. 1A). Using methylation-sensitive restriction digestion, we have determined that the region neighboring rs1621902 is methylated stronger on the paternal chr16q24.1 (Fig. S9).
Previously, we have proposed that FOXF1 locus may be responsible for key features of the maternal uniparental disomy 16, UPD(16), phenotype.5 In contrast to phenotypically benign paternal UPD(16), patients with maternal UPD(16) sometimes manifest features observed in ACDMPV, including heart defects, pulmonary arterial hypertension, tracheoesophageal fistula, gut malrotation, absent gallbladder, renal agenesis, hydronephrosis, imperforate anus, and single umbilical artery.1 This suggests that the stronger paternal enhancer may also act more ubiquitously than its maternal allele which may be more lung-specific. Corroboratively, the full lethal ACDMPV phenotype associated with CNV deletions on paternal chr16q24.1 in patients 122.3 and 204.3 and milder ACDMPV phenotype in the longer surviving patient 99.3 (who had lung transplantation) with a larger-sized CNV deletion on maternal chr16q24.1 (Fig. 1, S1) suggest that Unit 1 may act as a stronger lung enhancer on paternal chr16q24.1. According to the “seesaw” mechanism, higher methylation level of the 660UCR on paternal chr16q24.1 would favor binding of enhancerous EP300 through the exclusion of H3K4me3 methylases, whereas lower methylation of maternal allele of the LINC01082 regulatory interval in Unit 1 might increase expression of LINC01082 and thus suppress FOXF1. In contrast, the association of CNV deletion on maternal chromosome 16q24.1 in patient 144.3 (Fig. 1, S1) with full ACDMPV implies Unit 2 acting as a stronger lung enhancer on maternal chromosome 16q24.1.
In conclusion, we propose that two ultra-conserved intervals of the FOXF1 enhancer, 660UCR and 1000UCR, the latter overlapping the lncRNA genes LINC01082 and RP11-805I24.3, form Unit 1 that plays an essential role in human lung development. We propose that this unit acts as a stronger FOXF1/FENDRR enhancer on the paternal copy of chromosome 16q24.1 whereas Unit 2, with the typical chromatin epigenetic features of an active enhancer, may play a modifier role and act as a stronger lung enhancer on the maternal copy of chromosome 16q24.1. Moreover, the ultra-conserved non-coding enhancer intervals, especially those within Unit 1, can be traced down to lungfish, coelacanth (sarcopterygians) and even spotted gar (actinopterygians), suggesting that their appearance in fishes might have been one of important steps in lung evolution.
Author contributions
PSz and EYB, performed the molecular experiments, TM, TG, and JAK performed the computational analyzes, MB collected the clinical data, NC-S and GA evaluated the histopathological specimen, PSz, DW, and PSt analyzed and interpreted the molecular data, PSz and PSt designed the study concept, wrote, and critically revised the final version of the article.
Conflict of interests
The authors declare no conflicts of interest.
Funding
This work was supported by the grants awarded by the US National Institutes of Health (NIH), National Heart Lung and Blood Institute (NHLBI) R01HL137203 and Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD) grant R01HD087292 to P.St.
Acknowledgements
We thank Drs. E. Popek, L. Mayer, G. Huber, R. Mair, R. Gitter, G. Tulzer for helpful discussion. We thank Drs. I. Tiemann-Boege, T. Ebner, M. Witsch-Baumgartner, and G. Webersinke, and B. Poszewiecka for helpful discussion and technical assistance.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.05.002.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Stankiewicz P., Sen P., Bhatt S.S., et al. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet. 2009;84(6):780–791. doi: 10.1016/j.ajhg.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szafranski P., Dharmadhikari A.V., Brosens E., et al. Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder. Genome Res. 2013;23(1):23–33. doi: 10.1101/gr.141887.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slot E., Boers R., Boers J., et al. Genome wide DNA methylation analysis of alveolar capillary dysplasia lung tissue reveals aberrant methylation of genes involved in development including the FOXF1 locus. Clin Epigenetics. 2021;13(1):148. doi: 10.1186/s13148-021-01134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seo H., Kim J., Park G.H., Kim Y., Cho S.W. Long-range enhancers modulate Foxf1 transcription in blood vessels of pulmonary vascular network. Histochem Cell Biol. 2016;146(3):289–300. doi: 10.1007/s00418-016-1445-4. [DOI] [PubMed] [Google Scholar]
- 5.Schulze K.V., Szafranski P., Lesmana H., et al. Novel parent-of-origin-specific differentially methylated loci on chromosome 16. Clin Epigenetics. 2019;11(1):60. doi: 10.1186/s13148-019-0655-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.