Summary
Cytomegalovirus (CMV) infection remains one of the most frequent and life-threatening infectious complications after allogeneic hematopoietic stem cell transplantation (allo-HSCT). Herein, we comprehensively compared the immune cells of patients with uncontrolled and controlled CMV infection post-allo-HSCT and found that B-cells were extraordinarily insufficient because of impaired B-cells reconstitution in the uncontrolled infection group. Furthermore, in the controlled infection group, reconstructed B-cells showed signatures of mature B-cells, high expression of CXCR4 and IFITM1, and enrichment of CMV-associated B-cell receptors, which were lacking in the uncontrolled infection group. Consistently, sera from the uncontrolled infection group failed to inhibit CMV infection via neutralizing virus in vitro because of its lower content of anti-CMV-specific immunoglobulin G (IgG) than the controlled infection group. Overall, these results highlighted the contribution of B cells and anti-CMV-specific neutralizing IgGs to the restraint of CMV infection post-allo-HSCT, suggesting their potential as a supplementary treatment to improve outcomes.
Subject areas: Immunology, Virology, Stem cells research
Graphical abstract

Highlights
-
•
B-cells are insufficient in uncontrolled CMV infection patients after UCBT
-
•
CXCR4 and IFITM1 are decreased in B cells of uncontrolled infection patients
-
•
The enrichment of CMV-associated BCRs is lacking in uncontrolled infection patients
-
•
Prompt supplement of CMV-specific neutralizing antibodies inhibits CMV infection
Immunology; Virology; Stem cells research.
Introduction
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is used as a curative treatment for many hematologic malignancies, hematological disorders, and other immune deficiencies. However, many complications, such as acute graft-versus-host disease (aGVHD), pre-engraftment syndrome, and various pathogen infections, have hampered the improvement of prognosis (Einsele et al., 2020; Jin et al., 2021; Saberian et al., 2021). Cytomegalovirus (CMV) is a leading cause of post-transplant infections and was once called the troll of transplantation because of its high prevalence worldwide and its negative impact on the prognosis of allo-HSCT and solid organ transplant patients (Kumar and Selzner, 2020). Owing to the use of several direct anti-CMV agents, such as ganciclovir, valganciclovir, sodium phosphonate, and cidofovir, CMV reactivation, CMV disease, and mortality were significantly reduced among allo-HSCT recipients (Marty et al., 2017). However, prolonged and repeated administration of antiviral agents often leads to myelosuppression, unfavorable specific immune recovery, and drug resistance (El Chaer et al., 2016; Zamora et al., 2021). Moreover, long-term CMV infection has been associated with increased GVHD and no-relapse mortality, which are exacerbated by drug-induced myelosuppression (Akahoshi et al., 2021; El Chaer et al., 2016; El Helou and Razonable, 2019). Of interest, immunotherapy has revolutionized the management of a variety of diseases, especially cancer and infectious diseases (Arnaldez et al., 2020; Liu et al., 2021), and thus combining immunotherapy and antiviral drugs for long-term recurrent CMV infections after HSCT should be a promising therapeutic approach.
The application of adoptive CMV-specific T cell immunotherapy and vaccines has been attempted to mitigate the impact of infection and achieve improved therapeutic effects (Aldoss et al., 2020; Griffiths et al., 2011). However, these attempts encountered several challenges in early clinical studies, such as extending culture time in vitro, optimizing the purity of virus-specific T cell products, potential limitations of targeting a single viral antigen, and managing patients with uninfected donors (Roddie and Peggs, 2017). Moreover, several prophylaxis and treatment regimens for immunosuppression, mainly targeting T-cells, are widely applied for the prevention or attenuation of GVHD currently. Of note, adoptive transferred virus-specific T-cells or vaccinated cells might also be inhibited by immunosuppressants in vivo (Goldsmith et al., 2021; Liu et al., 2020). In addition, it is not fully understood whether other immune cells play important antiviral roles in the presence of immunosuppressants in patients after allo-HSCT. Thus, an analysis of the gene expression signature of the immune cell profile of patients using new high-throughput technology should be helpful in comprehensively evaluating the causes of failure to inhibit CMV infection in patients after allo-HSCT in the presence of immunosuppressants.
In this study, we comprehensively compared the composition of immune cells of patients with uncontrolled and controlled CMV infection using single-cell sequencing to clarify the cellular mechanisms responsible for improved prognosis. We found that the number of B-cells in patients with uncontrolled CMV infection was insufficient because of delayed reconstitution. Both CMV-associated B-cell antigen receptors (BCRs) and anti-CMV-specific immunoglobulin G (IgG) were deficient in patients with uncontrolled CMV infection. Importantly, our results indicated that exogenous supplementation of anti-CMV-specific IgG in patients with low specific IgG at 2 months after allo-HSCT effectively alleviated uncontrolled CMV infection, and could be used synergistically with antiviral agents to improve transplant outcomes.
Results
Uncontrolled CMV infection in patients after UCBT is associated with an unfavorable prognosis
First, we analyzed the cumulative incidence of CMV infection in 114 patients receiving unrelated cord blood transplantation (UCBT) and 82 patients receiving peripheral blood stem cell transplantation (PBSCT) within 240 days after transplantation (Figure S1 and Table S1). We found that although the incidence of infection in the PBSCT group was lower than that in the UCBT group (at day 240, 6⅛2 versus 101/114, p = 0.0126), it exceeded 70% in both groups (Figure 1A). To further assess the severity of CMV infection, we analyzed the duration of CMV activation in 170 patients with UCBT and 61 patients with PBSCT who experienced CMV infection (Figure 1B). We then divided patients into a controlled infection group (CMV reactivation duration <60 days) and an uncontrolled infection group (CMV reactivation duration ≥60 days or CMV disease accompanied by the death of the patient). Consistent with the trend of incidence, we observed that the proportion of uncontrolled infection in patients receiving UCBT was higher than that in those receiving PBSCT (57.64% versus 22.95%, p = 0.0001) (Figure 1C). These results suggested that UCBT is a suitable model for studying uncontrolled CMV infection after allo-HSCT.
Figure 1.
Association of uncontrolled CMV infection after allo-HSCT with an unfavorable prognosis
(A) Cumulative incidence of CMV infection in 114 cases with UCBT (purple line) and 82 cases with PBSCT (green line) for 240 days after allo-HSCT. Presentation of CMV DNAemia or CMV end-organ disease was defined as the occurrence of CMV infection. The data below the graph represent the number of patients without CMV infection at each indicated time.
(B) Distribution of percentages of the duration of CMV activation in 170 UCBT patients (top panel) and 61 PBSCT patients (bottom panel) with CMV DNA infection after allo-HSCT.
(C) Proportion of patients with controlled (CMV reactivation duration <60 days) and uncontrolled infection (CMV reactivation duration ≥60 days or CMV disease accompanied by the death of patient) in the UCBT (n = 170) and PBSCT (n = 61) groups.
(D) Overall survival of patients in the controlled (n = 70) and uncontrolled (n = 100) infection groups within 900 days after UCBT.
(E) Prognostic distribution in the uncontrolled and controlled infection groups 900 days after UCBT. Fisher’s exact tests were used in panels A (at day 240 after transplant), C (at day 240 after UCBT), and E (at day 900 after UCBT). Log-rank (Mantel-Cox) tests were performed in panel D. CMV, cytomegalovirus; allo-HSCT, allogeneic hematopoietic stem cell transplantation; UCBT, unrelated cord blood transplantation; PBSCT, peripheral blood stem cell transplantation.
To further determine the influence of uncontrolled CMV infection on long-term survival after allo-HSCT, we analyzed the survival of 170 patients with UCBT (Table S2). Our data showed that the survival of patients with uncontrolled CMV infection was significantly worse than that of patients with controlled infection within 30 months after transplantation (Figures 1D and 1E, p = 0.0358 and p = 0.0358, respectively). These data indicated that uncontrolled CMV infection after allo-HSCT is an important threat to transplant outcomes.
The number of B-cells was insufficient in patients with uncontrolled CMV infection after UCBT
To further clarify the role of immune subsets in controlling CMV infection after UCBT, we collected fresh PBMCs from 6 patients whose durations of CMV infection after transplantation were 28, 30, 55, 90, 92, and 120 days, respectively, for single-cell sequencing (Table S3). After removing low-quality cells, we obtained a total of 35,390 cells for further analysis (Figure S2A).
To systematically analyze immune cell populations, we normalized and summarized single-cell data from these 6 samples, and performed unsupervised clustering to identify distinguishable populations. We annotated these populations using their typical markers and successfully identified the major circulating immune cell types, including T-cells, NK cells, B-cells, monocytes, DCs, plasma cells, and a very small fraction (1.31%) of mixed classification cells. We noticed that the expression of classical markers for these cell types was consistent with the annotation (Figure S2B). We then divided patients into uncontrolled infection and controlled infection groups to compare the distribution of immune cells in the t-SNE plot between the 2 groups. As shown in Figures 2A and S2C, the most evident difference between the 2 groups was the insufficient number of B-cells in the uncontrolled infection group. Considering the important antiviral role of T-cells and NK cells, we analyzed the expression of related antiviral molecules between the 2 groups of patients. Our results showed that there was no significant difference in the expression of IFNG, TNF, PD1 (PDCD1), TIGIT, and NKG2D (KLRK1) between the two groups, despite the higher expression of KLRG1 in the controlled infection group (Figure S2D).
Figure 2.
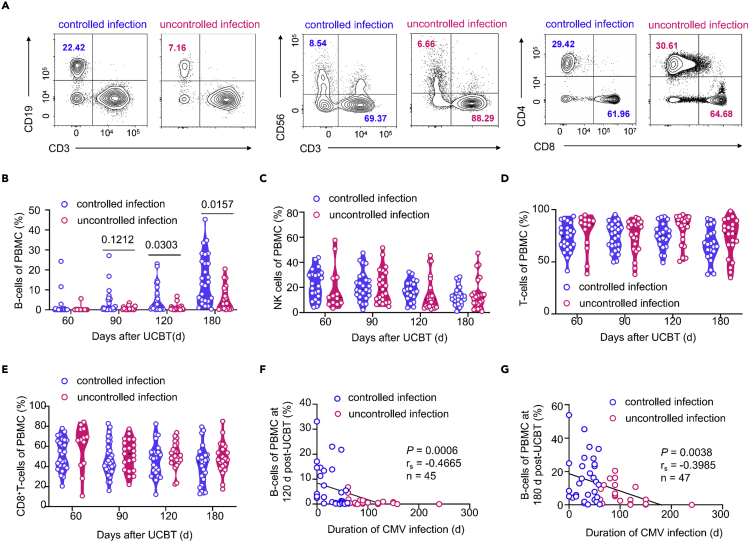
Insufficiency of B-cells in patients with uncontrolled CMV infection
(A) T-SNE plots of 35,900 high-quality peripheral blood mononuclear cells (PBMCs) from patients with controlled (n = 3) and uncontrolled (n = 3) infection showing the distribution of different cell populations based on the expression of known marker genes. PBMCs were collected from 6 patients 6 months after UCBT and then single-cell RNA sequencings were performed using the 10× Genomics platform. B-cells are highlighted by the dotted box.
(B) T-SNE plots of immune cell profiles of each patient. The duration of CMV infection of each patient is indicated by the depth of color.
(C) Fractions of B-cells, CD4+T-cells, CD8+T-cells, NK cells, monocytes, and dendritic cells in patients with controlled and uncontrolled infection. Each dot represented a patient. Data are shown as the mean ± SD and analyzed using an unpaired, two-tailed, Student’s t test.
Owing to the important antiviral role of T-cells and NK cells, we evaluated their ability to secrete antiviral cytokines in patients after UCBT (see Table S4 for patient characteristics). Surprisingly, we found that under the stimulation of PMA and ionomycin, the proportions of TNF-α+ CD8+T-cells, IFN-γ+ CD8+T-cells, TNF-α+ CD4+T-cells, and TNF-α+ NK cells were significantly lower than those in healthy controls (p<0.05, Figure S3), with no significant difference between the uncontrolled and controlled infection groups (p> 0.05, Figure S3), possibly because of the immunosuppressants received after transplantation. Moreover, we also stimulated PBMCs from patients baring alleles HLA-A∗0201 or HLA-A∗1101 with pp65-specific peptides and found that there was no significant difference in IFN-γ production between the controlled infection group (n = 10) and the uncontrolled infection group (n = 11), which were lower than the control group (Figure S4 and Table S5). These results suggested that the most important cause of the difference between controlled and uncontrolled infections might not be attributed to T and NK cells.
The distribution of immune cells in each patient is presented in Figure 2B. We detected that all 3 patients in the uncontrolled infection group had an anomalous reduction in the number of B-cells, especially pat#S6, whose CMV infection lasted for 120 days. In addition, there was no significant difference between the two groups in the proportion of other immune cells except B-cells (Figure 2C). These results indicated that the number of B-cells was insufficient in patients with uncontrolled CMV infection compared with the controlled CMV infection group.
B-cell reconstitution was impaired in patients with uncontrolled CMV infection after UCBT
To further confirm the difference in the content of B-cells between the uncontrolled and controlled infection groups, we dynamically monitored the immune cell subsets of patients at 60, 90, 120, and 180 days after transplantation (see Table S6 for patient characteristics). We found that the proportion of B-cells in the uncontrolled infection group was 0.5% (25th–75th quartile: 0.2–1), which was significantly lower than the 1.25% (0.3–11.83) observed in the controlled infection group on day 120 (p = 0.0303). Similar results were obtained on day 180 after transplantation (4.2% [0.5725–11.9] versus 12.81% [4.435–23.75], respectively, p = 0.0157, Figures 3A, 3B, and S5A). However, we did not detect any differences in the proportion and number of NK cells, T-cells, CD8+T-cells, or the ratio of CD4/CD8 at any time point (Figures 3A, 3C–3E, and S5, p> 0.05). Moreover, we noticed that the duration of CMV infection was negatively correlated with the proportion of B-cells on post-transplantation day 120 (p = 0.0006, r = −0.4665; Figure 3F) and 180 (p = 0.0038, r = −0.3985; Figure 3G). These data indicated that the reconstitution of B-cells is impaired in patients with uncontrolled CMV infection after transplantation.
Figure 3.
Impaired reconstitution of B-cells in patients with uncontrolled CMV infection
(A) Representative flow cytometry plots showing proportions of B-cells (CD3−CD19+), T-cells (CD3+CD56−), and NK cells (CD3−CD56+) in peripheral blood and proportions of CD4+T-cells, and CD8+T-cells in patients with controlled and uncontrolled infection at day 180 after UCBT.
(B–E) Violin plot showing proportions of B-cells (B), NK cells (C), T-cells (D), and CD8+T-cells (E) in the controlled infection group at day 60 (n= 27), 90 (n= 29),120 (n= 22), and 180 (n= 26) after UCBT, and uncontrolled infection group at day 60 (n= 13), 90 (n= 21),120 (n= 23), and 180 (n= 21) after UCBT. The proportion of cells between the two groups was analyzed using Mann-Whitney test.
(F) Correlations of proportions of peripheral blood B-cells in patients at day 120 (n = 45) and 180 (n = 47) (G) after UCBT with the duration of CMV infection (Pearson r correlation test).
Reconstituted B-cells are mature in patients after UCBT
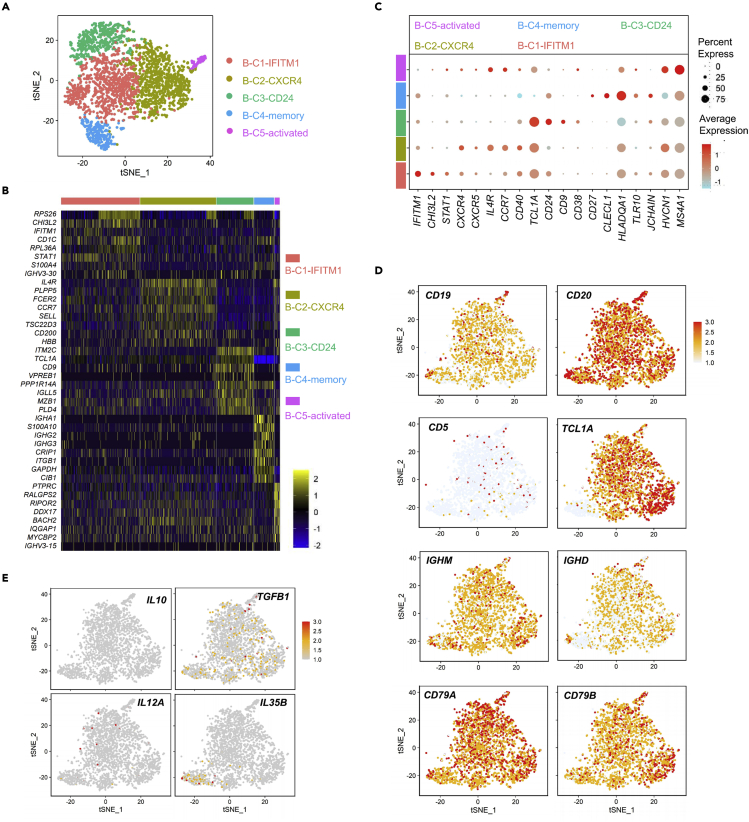
To further clarify the characteristics of reconstituted B-cells after transplantation, we performed an unsupervised clustering analysis using single-cell transcriptome data of 2938 B-cells from all 6 samples. Based on the differential expression of genes, we identified 5 distinguishable B-cell populations (Figures 4A and 4B), which were termed B-C1-IFITM1, B-C2-CXCR4, B-C3-CD24, B-C4-memory, and B-C5-activated according to their characteristic expressed genes (Figure 4C). We also analyzed the expression of several marker genes involved in B-cell maturation in these cell populations. We observed that CD19, CD20, TCL1A, IGHD, IGHM, CD79A, and CD79B were highly expressed in the cell population, whereas CD5 was low expressed, suggesting that the reconstituted B-cells were mature and exhibited a normal immune function (Figure 4D). Meanwhile, IL-10, TGFB1, IL12A, and IL35B (EBI3) were lowly expressed in these B-cells, indicating a reduced immune suppressive effect (Figure 4E). These results suggested that the reconstituted B-cells after UCBT are mature B-cells with antiviral potential.
Figure 4.
Maturation characteristics of reconstituted B-cells in patients after UCBT
(A) T-SNE plot of 2938 high-quality B-cells from 6 patients for subcluster visualization based on single-cell transcriptomes, with 5 clusters identified using unsupervised clustering which were termed B-C1-IFITM1, B-C2-CXCR4, B-C3-CD24, B-C4-memory, and B-C5-activated.
(B) Heatmap of top 8 cluster-specific gene expressions in each B-cell cluster. Gene expression is color-coded according to a scale based on Z score distribution.
(C) Dot plot of representative gene expressions in 5 B-cell subsets mentioned above, based on log2 normalized Z score (count +1).
(D) Same t-SNE plot as shown in panel (A), showing the expression of CD19, CD20, CD5, TCLA1, IGHM, IGHD, CD79A, and CD79B in each cluster.
(E) Same t-SNE plot as shown in panel (A), showing the expression of IL10, TGFB1, IL12A, and IL35B in B-cells. Low to high levels of expression are color-coded from gray to red.
CMV-associated BCRs are deficient in patients with uncontrolled CMV infection
To further determine whether the ability of antiviral B-cells to restrain CMV infection in patients with uncontrolled infection was impaired, we compared the distribution of B-cell subsets between uncontrolled and controlled infection groups. According to the t-SNE plot, compared with the controlled infection group, the B-C1-IFITM1and B-C2-CXCR4 populations highlighted by the dotted box were dramatically decreased in the uncontrolled infection group (Figure 5A). The CXCR4 signaling has been reported to direct Igκ recombination and orchestrate late B-cell lymphopoiesis (Mandal et al., 2019). In addition, interferon-inducible transmembrane protein 1 (IFITM1) restricts infection from enveloped DNA viruses that enter through the plasma membrane (Smith et al., 2019). The present study suggested that in patients with uncontrolled infection, the ability of B-cells to inhibit viral infection was compromised.
Figure 5.
Deficiency of CMV-associated BCRs in patients with uncontrolled CMV infection
(A) T-SNE plot of 2938 high-quality B-cells from patients with controlled and uncontrolled infection for subcluster visualization based on single-cell transcriptomes. The B-C1-IFITM1and B-C2-CXCR4 populations were highlighted by the dotted box.
(B) Heatmap of the expression of top 56 IGHV genes in 2938 B-cells. Gene expression is color-coded according to a scale based on Z score distribution. Genes that might be associated with CMV-specific recognition are marked in red.
(C) T-SNE plots of the expression of IGHV4-39, IGHV1-69D, IGHV4-34, and IGHV3-30 in B-cells from patients with controlled and uncontrolled infection. Low to high levels of expression are color-coded from gray to red.
(D) Dot plot of the expression of IGHV4-39, IGHV1-69D, IGHV4-34, and IGHV3-30 in patients with controlled and uncontrolled infection, based on log2 normalized Z score (count +1).
(E) RT-PCR analysis of the expression of IGHV4-39, IGHV1-69D, IGHV4-34, and IGHV3-30 in lymphocytes isolated from patients with controlled and uncontrolled infection (5 cases per group, day 60 after UCBT). ACTB was included as an internal control. Immunoglobulin heavy chain variable region, IGHV.
Given the essential role of specific BCR in recognizing viral antigens, we analyzed the expression of immunoglobulin heavy chain variable region (IGHV) genes in B-cells and detected the enrichment of certain IGHV genes (marked in red) that are associated with CMV recognition (Kostareli et al., 2009; Steininger et al., 2012) in the top 56 IGHV genes (Figure 5B). PBMCs from CMV seropositive donors were co-cultured with inactivated CMV, R848 (a TLR7/TLR8 agonist) and rhIL-2 for 7 days (Pinna et al., 2009), and a large number of plasma cells could be detected (Figures S6A and S6B). Enhanced expression of IGHV1-69D, IGHV3-30, IGHV4-34, and IGHV4-39 after stimulation were detected, although the dominant IGHV genes varied in different individuals (Figure S6C). Next, we compared the expression of IGHV4-39, IGHV1-69D, IGHV4-34, and IGHV3-30 between the 2 groups and found that IGHV4-39, IGHV1-69D, and IGHV4-34 were insufficient in B-cells from patients with uncontrolled infection (Figures 5C and 5D). Moreover, we verified the deficiency in the expression of IGHV4-39 and IGHV1-69D in some patients with uncontrolled infection whereas IGHV4-34 and IGHV3-30 were expressed in both groups (Figure 5E). These data suggested that B-cells in patients with uncontrolled CMV infection after UCBT were not only deficient in number but also lacked the expression of antiviral-related genes and the expansion of B-cell clones that specifically recognize the CMV antigen.
Prompt supplement of CMV-specific neutralizing antibodies facilitated inhibition of CMV infection
To further explore whether B-cells affect CMV infection by producing specific antibodies, we assessed the content of anti-CMV-specific immunoglobulin G (IgG) in the serum of 23 patients (Table S7) with CMV reactivation 2 months after UCBT (Figure S7). We found that the anti-CMV IgG content in the controlled infection group was higher compared with that in the uncontrolled infection group (247.2 [165.5–304.7] versus 142.3 [119.3–157.9] pg/mL, p = 0.0056; Figure 6A).
Figure 6.
Inhibition of CMV infection with serum from patients in the controlled infection group and CMV-specific neutralizing antibodies
(A) Violin plot showing concentrations of CMV-specific antibodies in serum of patients with controlled (n = 12) and uncontrolled infection (n = 11) at 2 months after UCBT using ELISA (Mann-Whitney test).
(B) Representative images of cytopathic effects of serum- or anti-CMV IgG-treated CMV on MRC-5 at 60 h after infection. Serum from patients in these 2 groups (n = 5) at 2 months after UCBT and anti-CMV IgG were diluted as indicated and incubated with CMV at 37°C for 30 min. Then, the neutralized CMV was used to infect MRC-5 cells. After 2 h, the supernatant was discarded and replaced with the medium containing 1% FBS for 48–72 h for further detection.
(C) Real-time PCR analysis of copies of the CMV IE1 gene in MRC-5 cells at 48 h after infection. Data are shown as the mean ± SD and analyzed using the unpaired, two-tailed Student’s t test.
(D and E) Western blotting analysis (D) and representative images of immunofluorescent staining (E) of the expression of IE1/2 and pp65 in MRC-5 cells 72 h after CMV infection (serum and anti-CMV IgG were diluted 1:16, and β-Actin was used as an internal control).
(F) Representative FACS plots (left panel) and statistical analysis (right panel) of the proportion of pp65+ MRC-5 cells 72 h after infection (serum and anti-CMV IgG were diluted 1:16, n = 5). Data are shown as the mean ± SD and analyzed using the unpaired, two-tailed Student’s t test. Each dot represented a patient. Dpi, days post infection.
Next, we applied the neutralization cell model in vitro to verify whether anti-CMV IgG in the post-transplant serum mediates neutralization and protection against CMV infection (Figure S7A, see Table S8 for patient characteristics). We noticed that at 16-fold dilution, serum from the controlled infection group and an exogenous high titer of anti-CMV IgG notably inhibited viral replication, whereas CMV was not neutralized by serum from the uncontrolled infection group, which showed the highest proliferation (Figure 6B). At a dilution of 1:64 or 1:256, only anti-CMV IgG effectively inhibited CMV replication, whereas the sera from both groups failed to attenuate proliferation (Figure 6B). We also observed that cytopathic effects (CPE) were most evident at 1-fold, 6-fold, and 25-fold dilutions in the uncontrolled infection group. However, anti-CMV IgG remarkably impeded CPE at 1:16, 1:64, and 1:256 dilutions, whereas CPE was inhibited at 16-fold dilution in the controlled infection group (Figure 6C). Confocal microscopy showed that almost all cells in the uncontrolled infection group expressed IE1/2 in the nucleus and pp65 in the cytoplasm, indicating that most cells were infected and underwent viral replication and synthesis (Figure 6D). In contrast, we observed a relatively scant expression of pp65 and IE1/2 in the other 2 groups, suggesting the remarkable inhibition of CMV infection. Western blotting and flow cytometry analyses further confirmed our findings (Figures 6E, 6F, and S7B). In addition, we found that anti-CMV IgG levels decreased significantly 6 months after UCBT when CMV DNaemia disappeared completely (Figure S7C).
To further explore the role of the addition of anti-CMV-specific IgG in the prevention of uncontrolled infection in vivo, we performed a retrospective analysis, including the control group (conventional anti-CMV prophylaxis, n = 64) and combination group (anti-CMV IgG and conventional prophylaxis). As shown in Table S9, the incidence of uncontrollable infection was reduced compared with the control group (33.75 versus 59.37%, p = 0.0025). There was no significant difference in the incidence of CMV between the two groups which may be related to more use of immunosuppressants (ruxolitinib and basiliximab) in the combination group because of the high incidence of aGVHD ≥2. These results indicated that CMV infection was not blocked in patients with uncontrolled infection because of an insufficient amount of CMV neutralizing antibody, whereas CMV infection might be effectively prevented by early and prompt anti-CMV IgG supplementation after UCBT.
Discussion
In this study, we showed that B-cells and anti-CMV-specific antibodies play a crucial role in restraining uncontrolled CMV infection after allo-HSCT. We focused on patients with uncontrolled CMV infection who experienced long-term CMV reactivation or death after allo-HSCT owing to poor overall survival (Reddehase, 2016). A moderate peak CMV titer has been associated with decreased recurrence, potentially because of the promotion of T cell reconstitution, whereas higher CMV titers, which correspond to the uncontrolled infection group in our study (Table S2), have been associated with higher non-relapse mortality (Admiraal et al., 2017; Leserer et al., 2021). Therefore, we proposed that the goal of CMV reactivation management is to prevent serious and prolonged infection while retaining controlled CMV reactivation to maintain enhanced T cell reconstitution and provide the graft-versus-leukemia effect. There were fewer stem cells from cord blood than from peripheral blood and bone marrow, leading to a greater risk of pathogen infection after transplantation (McGoldrick et al., 2013); we also noted that the longer durations of CMV infection coincided with UCBT patients in our study (Figure 1B). Therefore, we believe that UCBT is a suitable model for studying uncontrolled CMV infection after allo-HSCT.
Considerable evidence has suggested that NK cells and CMV-specific T-cells play a crucial role in protecting the virus from reactivation (El Haddad et al., 2019) and predicting the risk of CMV reactivation (Camargo et al., 2019). Studies in patients with PBSCT have shown that CMV-specific T cells from third-party sources have achieved similar efficacy to that from donors (Pei et al., 2017a, 2017b, 2022; Zhao et al., 2020). Because of donor limitation of UCBT, it is difficult to culture donor-derived virus-specific T cells, so third-party virus-specific T cell and γδ T-cells therapy may hold exciting prospects. In this study, we explored the antiviral function of T and NK cells. Nevertheless, the levels of cytokines secreted by T and NK cells in UCBT patients stimulated by ionomycin and PMA were significantly lower than those in healthy control patients, with no significant differences observed between the uncontrolled and controlled infection groups. It is worth mentioning that patients were treated with cyclosporine and mycophenolate mofetil after allo-HSCT, which might have affected the function of T and NK cells. Various factors contribute to the failure of CMV-specific T-cells to control CMV reactivation because of insufficient numbers in vivo (McGoldrick et al., 2013) CMV exposure has been reported to lead to impaired overall T-cell receptor diversity, but its impact on patient outcome requires further research (Suessmuth et al., 2015; Yeh et al., 2021). Consequently, an alternative anti-CMV mechanism might be involved in transplantation.
Cellular immunity, as well as innate and humoral immunity, is shown to play an important role in preventing CMV infection, but the exact mechanisms in different diseases are not well understood (Haidar et al., 2020). However, we provided evidence that the number of B-cells was insufficient in patients with uncontrolled CMV infection compared with that observed in the controlled infection group, because of their delayed reconstitution (Figures 2 and 3). CMV infection has been previously correlated with the proportion of highly mutated Ab genes (Wang et al., 2014). Reconstituted B-cells expressed various mature molecular markers and were enriched in many BCRs that might be related to CMV-specific recognition, indicating that B-cells are pivotal in CMV control. Furthermore, we found that CXCR4 and IFITM1 were highly expressed in B-cells from patients with controlled CMV infection. IFITM1, a member of the family of IFN-stimulated genes, prevents viral infection by preventing the entrance of the virus through the plasma membrane. CXCR4 signaling has been reported to guide the development of small pre-B and immature B-cells, including coordination of cell cycle exit, pre-BCR inhibition, IGK recombination, and BCR expression via activation of mitogen-activated protein kinase extracellular signal-regulated kinase (Becker et al., 2017; Mandal et al., 2019). Moreover, a previous study showed that the transfer of virus-specific memory B-cells provided long-term protection from lethal CMV infection in immunodeficient animals, suggesting its therapeutic potential (Klenovsek et al., 2007). However, further research is needed concerning the specific role and possible mechanism of IFITM1-and CXCR4-highexpression B-cells in resisting CMV infection.
Many patients experience damaged B-cell function or dependency on Ig substitution after allo-HSCT (van der Maas et al., 2019). Studies have shown that the levels of several antibodies, especially IgA and IgE, were significantly decreased within 180 days after allo-HSCT (Pei et al., 2017a, 2017b). Given that most patients received intravenous gamma globulin after allo-HSCT, it was difficult to accurately evaluate the IgG content generated by patients. Nevertheless, we discovered that the level of CMV-specific IgG in the controlled infection group was notably higher than that in patients with uncontrolled infection at 2 months after allo-HSCT, thus facilitating the inhibition of CMV infection by neutralizing the viral particles in vitro (Figure 6). These findings indicated that anti-CMV IgG could contribute to the protection from uncontrolled infection, consistent with the results from a murine model (Klenovsek et al., 2007; Martins et al., 2019). A recent study proposed that IgG binds to glycoprotein B present on the cell surface but not to the soluble antigen to mediate protective immunity (Jenks et al., 2020). In addition, anti-CMV IgG was shown to cooperate with γδ T-cells to contribute to the surveillance of CMV reactivation via the secretion of IFN-γ aggregated by IL12 and IFNα2 (Couzi et al., 2012). In our study, although sc-RNA seq showed an obvious proportion of γδ T-cells in the uncontrolled infection patients, it may fail to produce IFN-γ because of the effect of CsA, just like CD8+T cells. Of interest, we found that CMV was reactivated by day 20–30 after allo-HSCT in most patients, which might have been partially related to the 21-d half-life of plasma IgG (Morell et al., 1970). We believe that the neutralization titer of CMV antibodies is critical (exceeding 300 U/mL anti-CMV IgG neutralizing antibodies in our study), and might explain the differences in efficacy obtained in some clinical trials after allo-HSCT or organ transplant (Kornberg et al., 2020; Winston et al., 1982; Zamora et al., 2020; Zikos et al., 1998). Thanks to the breakthrough in protein spatial structure analysis and monoclonal antibody development in 2021, monoclonal antibodies are considered to have promising therapeutic prospects in the treatment and prevention of multiple viral infections, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), respiratory syncytial virus, and CMV (Thorp, 2021). Hence, the application of high titers of anti-CMV-specific neutralizing IgG to patients with low anti-CMV-specific IgG might be very helpful.
We also found that the amount of CMV-specific IgG was decreased soon after CMV antigen clearance (Figure S7C), suggesting that these antibodies might be derived from marginal zone B-cells and be short-lived, functioning as rapid-response antibodies to prevent the progression to uncontrolled infection. Consistent with this, extrafollicular B-cell responses might also play a key role in the humoral immune response to SARS-CoV-2, as anti-SARS-CoV-2 antibodies were found to decline within a few months after infection (Kaneko et al., 2020; Perreault et al., 2020). In this study, we provided evidence that the specific CMV IgG content at 2 months after allo-HSCT (about 1 month after first DNAemia) is of great significance in identifying patients at risk for uncontrolled infection.
In summary, persistent uncontrolled CMV infection after allo-HSCT remains an obstacle to improved prognosis. Our study highlighted the contribution of B-cells and anti-CMV-specific IgG in restraining uncontrolled CMV infection after allo-HSCT. We proposed that for patients with CMV reactivation, continuous monitoring of specific antibodies and BCR might be useful in identifying patients at high risk for progression to uncontrolled infection. For high-risk patients, timely supplementation with specific antibodies, potentially synergistically with cellular therapies, might effectively limit the degree of CMV infection.
Limitations of the study
This study had certain limitations. First, although the level of serum anti-CMV IgG around 2 months after allo-HSCT is important, because of the low number of peripheral B-cells in most patients at that time, it was difficult to select enough B-cells to identify their specific functional phenotype at the protein level. Second, because several IGHV genes examined above are broadly associated with antiviral immunity, more evidence is needed regarding CMV-specific humoral immunity, which will facilitate the development of high-affinity monoclonal antibodies. Third, we did not conduct a strict clinical randomized controlled trial to optimize the best prevention and treatment scheme for high-risk patients but rather proposed preventive anti-CMV IgG as a supplementary scheme. There is a possibility, however, that antibody therapy might not be satisfactory for patients whose CMV infection has persisted for a long time because of immune evasion from humoral immunity. CMV is known to escape immunity in certain ways, such as exploiting an ER-associated degradation pathway (Chandramouli et al., 2017), utilizing the Fc domain of the incorporated antibody to infect naive nonimmune cells, and encoding chemokines, cytokines, and chemokine receptors (Corrales-Aguilar et al., 2014; Manley et al., 2011). Thus, given the widespread application of antiviral drugs, future studies are needed to further define the strategy by which to combine antiviral agents with passive immunotherapy to achieve the best efficacy while avoiding immune evasion.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-CD45 | BD Biosciences | Cat# 563716; RRID:AB_2716864 |
| Mouse monoclonal anti-CD3 | Biolegend | Cat# 344836; RRID:AB_2565825 |
| Mouse monoclonal anti-CD56 | Biolegend | Cat# 318340; RRID:AB_2561944 |
| Mouse monoclonal anti-CD19 | Biolegend | Cat# 302217; RRID:AB_314247 |
| Mouse monoclonal anti-CD4 | Biolegend | Cat# 317410; RRID:AB_571955 |
| Mouse monoclonal anti-CD8a | Biolegend | Cat# 344750; RRID:AB_2687201 |
| Mouse monoclonal anti-IFN-γ | Biolegend | Cat# 506504; RRID:AB_315437 |
| Mouse monoclonal anti-TNF-α | BD Biosciences | Cat# 559321; RRID:AB_397219 |
| Mouse monoclonal anti-pp65 | Thermo Fisher Scientific | Cat# MA1-7296; RRID:AB_1016993 |
| Mouse monoclonal anti-IE1/2 | Abcam | Cat# ab53495; RRID:AB_882995 |
| Rabbit polyclonal anti-β-ACTIN | Proteintech | Cat# 20536-1-AP; RRID:AB_10700003 |
| Mouse monoclonal anti-CD138 | BioLegend | Cat# 356505; RRID:AB_2561879 |
| Goat Anti-Mouse IgG, HRP conjugate | Proteintech | Cat# SA00001-1; RRID:AB_2722565 |
| Goat Anti-Rabbit IgG(H + L), HRP conjugate | Proteintech | Cat# SA00001-2; RRID:AB_2722564 |
| Goat Anti-Mouse IgG, Cy3 conjugate | Proteintech | Cat# SA00009-1; RRID:AB_2814746 |
| Bacterial and virus strains | ||
| HCMV-AD169 | ATCC | ATCC VR-538 |
| Biological samples | ||
| Patient Blood Samples | The First Affiliated Hospital of USTC | http://www.ahslyy.com.cn/ |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco’s modified eagle’s medium | Gibco | Cat# 11995065 |
| Recombinant human IL-2 | Proteintech | Cat# HZ-1015 |
| FBS | Gibco | Cat# 10091-148 |
| R848 | Invivogen | Cat# tlrl-r848 |
| Ionomycin | Calbiochem | Cat# I3909 |
| TRIzol | Invitrogen | Cat# 15596018 |
| phorbol 12-myristate 13-acetate (PMA) | Sigma | Cat# P8139 |
| Monensin | Invitrogen | Cat# 00-4505-51 |
| Human CMV pp65 (495-503), NLVPMVATV | Sangon Biotech | Cat# T510162-0001 |
| Human CMV pp65 (501-509), ATVQGQNLK | Sangon Biotech | N/A |
| anti-CMV IgG, PH4 | Taibang Biologic Group | N/A |
| Critical commercial assays | ||
| anti-CMV IgG ELISA | Mei Mian Biotechnology | Cat# MM-13958H1 |
| Human Cytomegalovirus (HCMV) nucleic acid Detection Kit | DAAN GENE | Cat# 20143402167 |
| eBioscience Foxp3/Transcription Factor Staining Buffer Set | Thermo Fisher | Cat# 00-5523-00 |
| ProlongTM Diamond Antifade with DAPI | Invitrogen | Cat# P36941 |
| Premix Taq™PCR Reagents | TaKaRa | Cat# RR901A |
| PrimeScript™ IV 1st strand cDNA Synthesis Mix | TaKaRa | Cat# 6215A |
| TB Green® Premix Ex Taq™ II | TaKaRa | Cat# RR820W |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE192721 |
| Experimental models: Cell lines | ||
| MRC-5 | ATCC | ATCC CCL-171 RRID:CVCL_0440 |
| Oligonucleotides | ||
| Forward primer, IGHV4-34: 5′-TGTCTATGGTGGGTCCTTCAGTGG-3′ |
This paper | N/A |
| Reverse primer, IGHV4-34: 5′-GACGGGTTGTAGTTGGTGCTTCC-3′ |
This paper | N/A |
| Forward primer, IGHV3-30: 5′-GTGGGTGGCAGTTATATCA-3′ |
This paper | N/A |
| Reverse primer, IGHV3-30: 5′-GCTCTCAGGCTGTTCATT-3′ |
This paper | N/A |
| Forward primer, IGHV1-69D: 5′-CTTCTGGAGGCACCTTCA-3′ |
This paper | N/A |
| Reverse primer, IGHV1-69D: 5′-CTGTACCAAAGATAGGGATGAT-3′ |
This paper | N/A |
| Forward primer, IGHV4-39: 5′-TGGTGGCTCCATCAGCAGTAGTAG-3′ |
This paper | N/A |
| Reverse primer, IGHV4-39: 5′-TTGAGGGACGGGTTGTAGTAGGTG-3′ |
This paper | N/A |
| Forward primer, ACTB: 5′-AAGAGAGGCATCCTCACCCT-3′ |
(Wang et al., 2015) | N/A |
| Reverse primer, ACTB: 5′-TACATGGCTGGGGTGTTGAA-3′ |
(Wang et al., 2015) | N/A |
| Software and algorithms | ||
| GrapgPad Prism 9 | GraphPad | http://www.graphpad.com/, SCR_00279 |
| FlowJo_V10 | BD | https://www.flowjo.com/solutions/flowjo, SCR_008520 |
| R software (version 3.6.3) | r-project.org | https://www.r-project.org/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Haiming Wei (ustcwhm@ustc.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Ethics approval
This research received institutional approval from the Ethics Committee of the First Affiliated Hospital of the University of Science and Technology of China. All studies were performed following the Declaration of Helsinki.
Patients
This study evaluated CMV infection in patients at high risk for or who had refractory hematological malignancies and underwent allo-HSCT between January 2017 and April 2019. A total of 265 patients (137 males and 128 females, with an average age of 24.15) with complete clinical data who received a myeloablative conditioning regimen and a single unrelated cord blood unit (n = 183) or allogeneic peripheral blood stem cells (n = 82) were included in the analysis (Figure S1). Information on age, sex, and disease of patients were listed in Tables S1 and S2. Intravenous G-CSF (figeristine, 5–7 μg/kg/d) was administered from day +6 and continued until the absolute number of neutrophils exceeded 2.0 × 109/L for 3 consecutive days.
Surveillance and treatment of CMV infection
In the first 90 days following allo-HSCT, patients underwent twice-weekly CMV DNA surveillance via a real-time PCR (PCR) assay (DAAN GENE, Guangzhou, China). After post-allo-HSCT day 90, CMV surveillance was performed once a week until 2 consecutive tests were negative (Lodding et al., 2015). Sequencing of pathogenic microorganisms in the bronchial lavage fluid or urine was performed in some cases. Patients whose CMV DNA exceeded 1000 copies/mL after allo-HSCT were treated with ganciclovir or valganciclovir according to institutional standards.
Neutralization experiments
MRC-5 cells were cultured in DMEM medium (Gibco) supplemented with 10% FBS (Gibco) and penicillin/streptomycin at 37°C in a humidified atmosphere of 5% CO2 (Thermo Fisher Scientific). The CMV (AD169, MOI = 0.1), neutralized by serum or anti-CMV IgG (PH4, Taibang Biologic Group, China) at 1:16, 1:64, or 1:256 dilution for 30 min at 37°C, was used to infect MRC-5 cells. After infection for 2 h, the medium of cells was replaced with fresh DMEM containing 1% FBS for 48–72 h. Cells were harvested 48 h after infection for the detection of CMV replication using PCR. For the detection of virus proteins, cells were harvested 72 h after infection.
Method details
Definitions
CMV infection was diagnosed according to criteria based on underlying CMV risk characteristics: CMV DNA ≥100 copies/mL in peripheral blood by CMV PCR assay (Hakki et al., 2021). CMV duration time was defined as an accumulation of CMV positive time.
Sample collection and preparation for single-cell sequencing
At 6-month after allo-HSCT, fresh peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation from fresh blood of 6 patients collected into EDTA-coated tubes (Table S3) after providing informed consent. Lysis buffer (cat. 555,899, BD Biosciences) was used to rapidly lyse a small amount of the remaining red blood cells. Cells that passed the test (cell survival rate >90%) were washed and resuspended in DMEM supplemented with 10% fetal bovine serum (FBS, Gibco) to prepare a suitable cell concentration of 800–1100 cells/μL for analysis using 10× Genomics Chromium.
RNA-seq library preparation for 10× genomics single-cell 5′ sequencing
Cells were loaded onto a 10× chromium single-cell platform (Single-Cell 3′ library and Gel Bead Kit v.3) and the generation of gel beads in emulsion (GEMs), barcoding, GEM-RT clean-up, cDNA amplification, and library construction were all performed according to the manufacturer’s instructions. Qubit was used for library quantification before pooling. The final library pool was sequenced using 150 bp paired-end reads in the Illumina NovaSeq 6000 system.
Data processing of single-cell RNA-seq
FASTQ files were processed using the 10× Genomics Cell Ranger (3.0.1 version) to map the GRCh38 reference genome and generate a matrix containing normalized gene counts versus cells per sample. All functions were performed using default arguments unless otherwise noted. Following the exclusion of low-quality cells (<200 genes or >8% mitochondrial genes/cells), 35,390 cells were included for downstream analyses. Datasets from different samples were integrated using Seurat v3 with default parameters to remove the batch effect.
Dimensionality reduction, clustering, and annotation
We used the Seurat function ‘FindVariableFeatures’ to identify highly variable genes (HVGs), and used the top 1000 HVGs for data integration after removing TRAV and TRBV genes to avoid interference of TCR gene bias in cluster analysis. Principal component analysis was performed; t-SNE was used for dimensionality reduction, with the top 30 principal components adopted for the ‘FindNeighbors’ function, while a resolution parameter of 0.5 was utilized for the ‘FindClusters’ function. Marker genes of each cell cluster were recognized by the ‘FindAllMarkers’ function, and the whole dataset was then categorized into T-cells, NK cells, monocytes, B-cells, dendritic cells (DCs), plasma cells, and other cells (including neutrophils and red cells) according to known markers. Subsequently, 2938 B-cells were further categorized into 5 clusters using a resolution parameter of 0.3.
Stimulation of PBMCs in vitro and staining
Freshly isolated PBMCs were resuspended and seeded in 24 well plates in RPMI 1640 medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and penicillin (100 U/mL)/streptomycin (100 μg/mL). Cells were stimulated with ionomycin (1μg/mL, Calbiochem) and phorbol 12-myristate 13-acetate (PMA) (50ng/mL, Sigma) or HLA-restricted CMV pp65 peptides (key resources table) in the presence of monensin (10μg/mL, Sigma) for 4hat 37°C and 5% CO2, as previously described (Wang et al., 2021). Stimulated cells were collected and stained with monoclonal antibodies (mAbs, key resources table) against CD45, CD3, CD56, and CD8, followed by permeabilization (eBioscience) for 20 min at 20°C, and then stained with mAbs (key resources table) against interferon-γ (IFN-γ) and tumor necrosis factor alpha (TNF-α) (Wang et al., 2019). Information on baseline Characteristics of patients were listed in Tables S4 and S5.
Peripheral lymphocyte detection
PBMCs from patients at day 60, 90, 120, and 180 after UCBT were isolated and stained with mAbs against CD45, CD3, CD56, CD19, CD4, and CD8 for flow cytometry.
Flow cytometry and analysis
Data were collected using a NovoCyte flow cytometer (Agilent, USA) and analyzed using the FlowJo software.
Immunoblotting
Infected cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 10% Tris-glycine gels. After transfer to polyvinylidene difluoride (PVDF) membranes, membranes were blocked with QuickBlock Western (Beyotime) for 15 min at 20°C and probed with the following mouse monoclonal antibodies: anti-IE1/2, anti-pp65, and anti-β-Actin (1:1000, Proteintech). After incubation with horseradish peroxidase-conjugated secondary antibodies (1:3000, Proteintech), protein bands were visualized by chemiluminescence autoradiography (Clinx, Shanghai, China).
Reverse transcription-PCR
Total RNA of PBMCs obtained from patients was extracted using the TRIzol reagent (Invitrogen) and then reverse-transcribed to cDNA (Takara) according to the manufacturer’s instructions. Touchdown PCR was performed using the Premix Taq kit (Takara) in a PCR System (Bio-Rad Laboratories). Reactions were amplified in a thermocycler at 96°C for 2 min, 65°C for 30 s, then 9 cycles of 96°C for 30 s, 65°C for 30 s, then 9 cycles of 96°C for 30 s, 62°C for 30 s, and then 9 cycles of 96°C for 30s, 59°C for 30 s, followed by 30 s at 72°C. The primers used are listed in the Key resources table.
Culture of CMV-associated memory plasma cells
Freshly isolated PBMCs from the healthy control group (CMV seropositive, n = 3, 1–2×106 cells/culture) were co-cultured with CMV inactivated, R848 (1μg/mL, InvivoGen) and recombinant human IL-2 (10 ng/mL, Peprotech) for 7 days in RPMI 1640 medium supplemented with 10% FBS at 37°C and 5% CO2. Half of the medium was changed every two days. Stimulated cells were collected and stained with APC-CD138 for flow cytometry. IGHV gene expression was detected by real-time PCR.
Enzyme-linked immunosorbent assay (ELISA)
Sera from patients collected 2 months after allo-HSCT was measured for anti-CMV IgG by ELISA according to the manufacturer’s guidelines (Mei Mian Biotechnology, Jiangsu, China).
Immunofluorescence staining and confocal microscopy
MRC-5 cells infected with CMV were fixed with 4% paraformaldehyde for 30 min at 20°C. After blocking with 5% goat serum (Beyotime) and permeabilization with 0.25% Triton X-100, cells were stained with anti-IE1/2 (1:100) at 4°C overnight on a rocker. Cells were then washed twice and labeled with goat anti-mouse IgG1-Cy3 secondary antibodies (1:300) for 30 min at 20°C. After staining with anti-pp65-FITC, cell slides were mounted on microscope slides using ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific). Images were captured under a confocal laser scanning microscope (LSM 880, Zeiss).
Quantification and statistical analysis
Several R packages (Seurat, ggplot2, pheatmap) were used for single-cell RNA-seq data manipulation and visualization. Statistical analyses were performed using GraphPad Prism version 9.0 (La Jolla, California, USA) with the appropriate tests outlined in figure legends unless otherwise noted. Each dot represented a patient. Differences in categorical clinical variates in patients were analyzed by Fisher exact test or χ2 test. Logrank (Mantel-Cox) tests were performed to compare the overall survival of patients. Unpaired Student t-tests or Mann-Whitney tests were performed to analyze differences between 2 independent groups. One-way ANOVA was used to analyze the data comparison between more than two groups. Pearson r correlation test was used to analyze the relationship between B-cells proportion and duration of CMV infection. The values of p< 0.05 were considered statistically significant.
Acknowledgments
We acknowledge Prof. Shenghai Huang (Anhui Medical University, Hefei, China) and Prof. Jing An (Capital Medical University, Beijing, China) for their generous help with experimental materials. We also would like to thank Editage (www.editage.com) for English language editing. The single-cell RNA-seq was supported by Genergy Biotechnology of Shanghai. This study was supported by the National Natural Science Foundation of China (81903637, 82100230), the Key Projects of Research and Development Program of Anhui Province (201904a07020094), and the Fundamental Research Funds for the Central Universities (WK9110000001, WK9110000027, WK9110000168) in China.
Author contributions
H.R.W. performed experimental work and analyzed and interpreted the data. H.L.L. and B.L.W. provided study materials or patients, supervised the research, and interpreted the data. L.Z., S.S.W., Y.W., and M.J.T. provided study materials or patients. Q.L., Z.M.S., D.Y.W., X.H.Z., and B.Q.F. provided strategic planning and advice. H.M.W. designed experiments, supervised the research, and analyzed and interpreted the data.
Declaration of interests
The authors declare that they have no conflict of interest.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105065.
Contributor Information
Huilan Liu, Email: huilanl@ustc.edu.cn.
Baolong Wang, Email: wbl196555@ustc.edu.cn.
Haiming Wei, Email: ustcwhm@ustc.edu.cn.
Supplemental information
Data and code availability
Original single-cell RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus repository under accession number: GSE192721.
This paper does not report the original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Admiraal R., de Koning C.C.H., Lindemans C.A., Bierings M.B., Wensing A.M.J., Versluys A.B., Wolfs T.F.W., Nierkens S., Boelens J.J. Viral reactivations and associated outcomes in the context of immune reconstitution after pediatric hematopoietic cell transplantation. J. Allergy Clin. Immunol. 2017;140:1643–1650.e9. doi: 10.1016/j.jaci.2016.12.992. [DOI] [PubMed] [Google Scholar]
- Akahoshi Y., Kimura S.I., Inamoto Y., Seo S., Muranushi H., Shimizu H., Ozawa Y., Tanaka M., Uchida N., Kanda Y., et al. Effect of cytomegalovirus reactivation with or without acute graft-versus-host disease on the risk of nonrelapse mortality. Clin. Infect. Dis. 2021;73:e620–e628. doi: 10.1093/cid/ciaa1871. [DOI] [PubMed] [Google Scholar]
- Aldoss I., La Rosa C., Baden L.R., Longmate J., Ariza-Heredia E.J., Rida W.N., Lingaraju C.R., Zhou Q., Martinez J., Kaltcheva T., et al. Poxvirus vectored cytomegalovirus vaccine to prevent cytomegalovirus viremia in transplant recipients: a Phase 2, randomized clinical trial. Ann. Intern. Med. 2020;172:306–316. doi: 10.7326/M19-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaldez F.I., O’Day S.J., Drake C.G., Fox B.A., Fu B., Urba W.J., Montesarchio V., Weber J.S., Wei H., Wigginton J.M., Ascierto P.A. The Society for Immunotherapy of Cancer perspective on regulation of interleukin-6 signaling in COVID-19-related systemic inflammatory response. J. Immunother. Cancer. 2020;8:e000930. doi: 10.1136/jitc-2020-000930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M., Hobeika E., Jumaa H., Reth M., Maity P.C. CXCR4 signaling and function require the expression of the IgD-class B-cell antigen receptor. Proc. Natl. Acad. Sci. USA. 2017;114:5231–5236. doi: 10.1073/pnas.1621512114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo J.F., Wieder E.D., Kimble E., Benjamin C.L., Kolonias D.S., Kwon D., Chen X.S., Komanduri K.V. Deep functional immunophenotyping predicts risk of cytomegalovirus reactivation after hematopoietic cell transplantation. Blood. 2019;133:867–877. doi: 10.1182/blood-2018-10-878918. [DOI] [PubMed] [Google Scholar]
- Chandramouli S., Malito E., Nguyen T., Luisi K., Donnarumma D., Xing Y., Norais N., Yu D., Carfi A. Structural basis for potent antibody-mediated neutralization of human cytomegalovirus. Sci. Immunol. 2017;2:eaan1457. doi: 10.1126/sciimmunol.aan1457. [DOI] [PubMed] [Google Scholar]
- Corrales-Aguilar E., Trilling M., Hunold K., Fiedler M., Le V.T., Reinhard H., Ehrhardt K., Mercé-Maldonado E., Aliyev E., Zimmermann A., et al. Human Cytomegalovirus Fcgamma binding proteins gp34 and gp68 antagonize Fcgamma receptors I, II and III. PLoS Pathog. 2014;10:e1004131. doi: 10.1371/journal.ppat.1004131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzi L., Pitard V., Sicard X., Garrigue I., Hawchar O., Merville P., Moreau J.F., Déchanet-Merville J. Antibody-dependent anti-cytomegalovirus activity of human γδ T cells expressing CD16 (FcγRIIIa) Blood. 2012;119:1418–1427. doi: 10.1182/blood-2011-06-363655. [DOI] [PubMed] [Google Scholar]
- Einsele H., Ljungman P., Boeckh M. How I treat CMV reactivation after allogeneic hematopoietic stem cell transplantation. Blood. 2020;135:1619–1629. doi: 10.1182/blood.2019000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Chaer F., Shah D.P., Chemaly R.F. How I treat resistant cytomegalovirus infection in hematopoietic cell transplantation recipients. Blood. 2016;128:2624–2636. doi: 10.1182/blood-2016-06-688432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Haddad L., Ariza-Heredia E., Shah D.P., Jiang Y., Blanchard T., Ghantoji S.S., El Chaer F., El-Haddad D., Prayag A., Nesher L., et al. The ability of a cytomegalovirus ELISPOT assay to predict outcome of low-level CMV reactivation in hematopoietic cell transplant recipients. J. Infect. Dis. 2019;219:898–907. doi: 10.1093/infdis/jiy592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Helou G., Razonable R.R. Letermovir for the prevention of cytomegalovirus infection and disease in transplant recipients: an evidence-based review. Infect. Drug Resist. 2019;12:1481–1491. doi: 10.2147/IDR.S180908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith S.R., Abid M.B., Auletta J.J., Bashey A., Beitinjaneh A., Castillo P., Chemaly R.F., Chen M., Ciurea S., Dandoy C.E., et al. Posttransplant cyclophosphamide is associated with increased cytomegalovirus infection: a CIBMTR analysis. Blood. 2021;137:3291–3305. doi: 10.1182/blood.2020009362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths P.D., Stanton A., McCarrell E., Smith C., Osman M., Harber M., Davenport A., Jones G., Wheeler D.C., O’Beirne J., et al. Cytomegalovirus glycoprotein-B vaccine with MF59 adjuvant in transplant recipients: a phase 2 randomised placebo-controlled trial. Lancet. 2011;377:1256–1263. doi: 10.1016/S0140-6736(11)60136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidar G., Boeckh M., Singh N. Cytomegalovirus infection in solid organ and hematopoietic cell transplantation: state of the evidence. J. Infect. Dis. 2020;221:S23–S31. doi: 10.1093/infdis/jiz454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakki M., Aitken S.L., Danziger-Isakov L., Michaels M.G., Carpenter P.A., Chemaly R.F., Papanicolaou G.A., Boeckh M., Marty F.M. American society for transplantation and cellular therapy series: # 3-prevention of cytomegalovirus infection and disease after hematopoietic cell transplantation. Transpl.Cell Ther. 2021;27:707–719. doi: 10.1016/j.jtct.2021.05.001. [DOI] [PubMed] [Google Scholar]
- Jenks J.A., Nelson C.S., Roark H.K., Goodwin M.L., Pass R.F., Bernstein D.I., Walter E.B., Edwards K.M., Wang D., Fu T.M., et al. Antibody binding to native cytomegalovirus glycoprotein B predicts efficacy of the gB/MF59 vaccine in humans. Sci. Transl. Med. 2020;12:eabb3611. doi: 10.1126/scitranslmed.abb3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Sun Z., Liu H., Zhu X., Zhou Y., Fu B., Zheng X., Song K., Tang B., Wu Y., et al. Inflammatory monocytes promote pre-engraftment syndrome and tocilizumab can therapeutically limit pathology in patients. Nat. Commun. 2021;12:4137. doi: 10.1038/s41467-021-24412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko N., Kuo H.H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., Piechocka-Trocha A., Lefteri K., Osborn M., Bals J., et al. Loss of Bcl-6-Expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020;183:143–157.e13. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenovsek K., Weisel F., Schneider A., Appelt U., Jonjic S., Messerle M., Bradel-Tretheway B., Winkler T.H., Mach M. Protection from CMV infection in immunodeficient hosts by adoptive transfer of memory B cells. Blood. 2007;110:3472–3479. doi: 10.1182/blood-2007-06-095414. [DOI] [PubMed] [Google Scholar]
- Kornberg A., Witt U., Kornberg J., Müller K., Friess H., Thrum K. Prophylactic anti-cytomegalovirus hyperimmunoglobulin in critically ill liver transplant patients: impact on early immunology and survival. J. Clin. Med. 2020;9:656. doi: 10.3390/jcm9030656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostareli E., Hadzidimitriou A., Stavroyianni N., Darzentas N., Athanasiadou A., Gounari M., Bikos V., Agathagelidis A., Touloumenidou T., Zorbas I., et al. Molecular evidence for EBV and CMV persistence in a subset of patients with chronic lymphocytic leukemia expressing stereotyped IGHV4-34 B-cell receptors. Leukemia. 2009;23:919–924. doi: 10.1038/leu.2008.379. [DOI] [PubMed] [Google Scholar]
- Kumar D., Selzner N. Cytomegalovirus: the ‘troll of transplantation’ is now the ‘troll of tolerance. Transplantation. 2020;104:238–239. doi: 10.1097/TP.0000000000002894. [DOI] [PubMed] [Google Scholar]
- Leserer S., Bayraktar E., Trilling M., Bogdanov R., Arrieta-Bolaños E., Tsachakis-Mück N., Crivello P., Koldehoff M., Maaßen F., Ross R.S., et al. Cytomegalovirus kinetics after hematopoietic cell transplantation reveal peak titers with differential impact on mortality, relapse and immune reconstitution. Am. J. Hematol. 2021;96:436–445. doi: 10.1002/ajh.26094. [DOI] [PubMed] [Google Scholar]
- Liu J., Gao H., Xu L.P., Mo X.D., Liu R., Liang S., Wu N., Wang M., Wang Z., Chang Y.J., et al. Immunosuppressant indulges EBV reactivation and related lymphoproliferative disease by inhibiting Vdelta2+ T cells activities after hematopoietic transplantation for blood malignancies. J. Immunother. Cancer. 2020;8:e000208. doi: 10.1136/jitc-2019-000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Hogg G.D., DeNardo D.G. Rethinking immune checkpoint blockade: ‘Beyond the T cell’. J. Immunother. Cancer. 2021;9:e001460. doi: 10.1136/jitc-2020-001460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodding I.P., Sengeløv H., da Cunha-Bang C.C., Iversen M., Rasmussen A., Gustafsson F., Downing J.G., Grarup J., Kirkby N., Frederiksen C.M., et al. Clinical application of variation in replication kinetics during episodes of post-transplant cytomegalovirus infections. EBioMedicine. 2015;2:699–705. doi: 10.1016/j.ebiom.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M., Okoreeh M.K., Kennedy D.E., Maienschein-Cline M., Ai J., McLean K.C., Kaverina N., Veselits M., Aifantis I., Gounari F., Clark M.R. CXCR4 signaling directs Igk recombination and the molecular mechanisms of late B lymphopoiesis. Nat. Immunol. 2019;20:1393–1403. doi: 10.1038/s41590-019-0468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley K., Anderson J., Yang F., Szustakowski J., Oakeley E.J., Compton T., Feire A.L. Human cytomegalovirus escapes a naturally occurring neutralizing antibody by incorporating it into assembling virions. Cell Host Microbe. 2011;10:197–209. doi: 10.1016/j.chom.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Martins J.P., Andoniou C.E., Fleming P., Kuns R.D., Schuster I.S., Voigt V., Daly S., Varelias A., Tey S.K., Degli-Esposti M.A., Hill G.R. Strain-specific antibody therapy prevents cytomegalovirus reactivation after transplantation. Science. 2019;363:288–293. doi: 10.1126/science.aat0066. [DOI] [PubMed] [Google Scholar]
- Marty F.M., Ljungman P., Chemaly R.F., Maertens J., Dadwal S.S., Duarte R.F., Haider S., Ullmann A.J., Katayama Y., Brown J., et al. Letermovir prophylaxis for cytomegalovirus in hematopoietic-cell transplantation. N. Engl. J. Med. 2017;377:2433–2444. doi: 10.1056/NEJMoa1706640. [DOI] [PubMed] [Google Scholar]
- McGoldrick S.M., Bleakley M.E., Guerrero A., Turtle C.J., Yamamoto T.N., Pereira S.E., Delaney C.S., Riddell S.R. Cytomegalovirus-specific T cells are primed early after cord blood transplant but fail to control virus in vivo. Blood. 2013;121:2796–2803. doi: 10.1182/blood-2012-09-453720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell A., Terry W.D., Waldmann T.A. Metabolic properties of IgG subclasses in man. J. Clin. Invest. 1970;49:673–680. doi: 10.1172/JCI106279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei X.Y., Liu X.F., Zhao X.Y., Lv M., Mo X.D., Chang Y.J., Shang Q.N., Sun Y.Q., Chen Y.H., Xu L.P., et al. Comparable anti-CMV responses of transplant donor and third-party CMV-specific T cells for treatment of CMV infection after allogeneic stem cell transplantation. Cell. Mol. Immunol. 2022;19:482–491. doi: 10.1038/s41423-021-00829-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei X.Y., Zhao X.Y., Chang Y.J., Liu J., Xu L.P., Wang Y., Zhang X.H., Han W., Chen Y.H., Huang X.J. Cytomegalovirus-specific T-cell transfer for refractory cytomegalovirus infection after haploidentical stem cell transplantation: the quantitative and qualitative immune recovery for cytomegalovirus. J. Infect. Dis. 2017;216:945–956. doi: 10.1093/infdis/jix357. [DOI] [PubMed] [Google Scholar]
- Pei X.Y., Zhao X.Y., Xu L.P., Wang Y., Zhang X.H., Chang Y.J., Huang X.J. Immune reconstitution in patients with acquired severe aplastic anemia after haploidentical stem cell transplantation. Bone Marrow Transplant. 2017;52:1556–1562. doi: 10.1038/bmt.2017.174. [DOI] [PubMed] [Google Scholar]
- Perreault J., Tremblay T., Fournier M.J., Drouin M., Beaudoin-Bussières G., Prévost J., Lewin A., Bégin P., Finzi A., Bazin R. Waning of SARS-CoV-2 RBD antibodies in longitudinal convalescent plasma samples within 4 months after symptom onset. Blood. 2020;136:2588–2591. doi: 10.1182/blood.2020008367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna D., Corti D., Jarrossay D., Sallusto F., Lanzavecchia A. Clonal dissection of the human memory B-cell repertoire following infection and vaccination. Eur. J. Immunol. 2009;39:1260–1270. doi: 10.1002/eji.200839129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddehase M.J. Mutual interference between cytomegalovirus and reconstitution of protective immunity after hematopoietic cell transplantation. Front. Immunol. 2016;7:294. doi: 10.3389/fimmu.2016.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roddie C., Peggs K.S. Immunotherapy for transplantation-associated viral infections. J. Clin. Invest. 2017;127:2513–2522. doi: 10.1172/JCI90599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saberian C., Abdel-Wahab N., Abudayyeh A., Rafei H., Joseph J., Rondon G., Whited L., Gruschkus S., Fa’Ak F., Daher M., et al. Post-transplantation cyclophosphamide reduces the incidence of acute graft-versus-host disease in patients with acute myeloid leukemia/myelodysplastic syndromes who receive immune checkpoint inhibitors after allogeneic hematopoietic stem cell transplantation. J. Immunother. Cancer. 2021;9:e001818. doi: 10.1136/jitc-2020-001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.E., Busse D.C., Binter S., Weston S., Diaz Soria C., Laksono B.M., Clare S., Van Nieuwkoop S., Van den Hoogen B.G., Clement M., et al. Interferon-induced transmembrane Protein 1 restricts replication of viruses that enter cells via the plasma membrane. J. Virol. 2019;93:e02003–e02018. doi: 10.1128/JVI.02003-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steininger C., Widhopf G.F., Ghia E.M., Morello C.S., Vanura K., Sanders R., Spector D., Guiney D., Jäger U., Kipps T.J. Recombinant antibodies encoded by IGHV1-69 react with pUL32, a phosphoprotein of cytomegalovirus and B-cell superantigen. Blood. 2012;119:2293–2301. doi: 10.1182/blood-2011-08-374058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suessmuth Y., Mukherjee R., Watkins B., Koura D.T., Finstermeier K., Desmarais C., Stempora L., Horan J.T., Langston A., Qayed M., et al. CMV reactivation drives posttransplant T-cell reconstitution and results in defects in the underlying TCRβ repertoire. Blood. 2015;125:3835–3850. doi: 10.1182/blood-2015-03-631853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorp H.H. Proteins, proteins everywhere. Science. 2021;374:1415. doi: 10.1126/science.abn5795. [DOI] [PubMed] [Google Scholar]
- van der Maas N.G., Berghuis D., van der Burg M., Lankester A.C. B cell reconstitution and influencing factors after hematopoietic stem cell transplantation in children. Front. Immunol. 2019;10:782. doi: 10.3389/fimmu.2019.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu Y., Xu L.T., Jackson K.J., Roskin K.M., Pham T.D., Laserson J., Marshall E.L., Seo K., Lee J.Y., et al. Effects of aging, cytomegalovirus infection, and EBV infection on human B cell repertoires. J. Immunol. 2014;192:603–611. doi: 10.4049/jimmunol.1301384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fu B., Shen X., Guo C., Liu Y., Zhang J., Sun R., Ye Y., Li J., Tian Z., Wei H. Restoration of HBV-specific CD8+ T-cell responses by sequential low-dose IL-2 treatment in non-responder patients after IFN-α therapy. Signal Transduct. Target.Ther. 2021;6:376. doi: 10.1038/s41392-021-00776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zheng X., Fu B., Nian Z., Qian Y., Sun R., Tian Z., Wei H. Hepatectomy promotes recurrence of liver cancer by enhancing IL-11-STAT3 signaling. EBioMedicine. 2019;46:119–132. doi: 10.1016/j.ebiom.2019.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Sun Q., Wu Y., Wang L., Zhou C., Ma W., Zhang Y., Wang S., Zhang S. Granzyme M expressed by tumor cells promotes chemoresistance and EMT in vitro and metastasis in vivo associated with STAT3 activation. Oncotarget. 2015;6:5818–5831. doi: 10.18632/oncotarget.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston D.J., Pollard R.B., Ho W.G., Gallagher J.G., Rasmussen L.E., Huang S.N., Lin C.H., Gossett T.G., Merigan T.C., Gale R.P. Cytomegalovirus immune plasma in bone marrow transplant recipients. Ann. Intern. Med. 1982;97:11–18. doi: 10.7326/0003-4819-97-1-11. [DOI] [PubMed] [Google Scholar]
- Yeh A.C., Varelias A., Reddy A., Barone S.M., Olver S.D., Chilson K., Onstad L.E., Ensbey K.S., Henden A.S., Samson L., et al. CMV exposure drives long-term CD57+ CD4 memory T-cell inflation following allogeneic stem cell transplant. Blood. 2021;138:2874–2885. doi: 10.1182/blood.2020009492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora D., Duke E.R., Xie H., Edmison B.C., Akoto B., Kiener R., Stevens-Ayers T., Wagner R., Mielcarek M., Leisenring W.M., et al. Cytomegalovirus-specific T-cell reconstitution following letermovir prophylaxis after hematopoietic cell transplantation. Blood. 2021;138:34–43. doi: 10.1182/blood.2020009396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora D., Krantz E.M., Green M.L., Joncas-Schronce L., Blazevic R., Edmison B.C., Huang M.L., Stevens-Ayers T., Jerome K.R., Geballe A.P., Boeckh M. Cytomegalovirus humoral response against epithelial cell entry-mediated infection in the primary infection setting after hematopoietic cell transplantation. J. Infect. Dis. 2020;221:1470–1479. doi: 10.1093/infdis/jiz596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.Y., Pei X.Y., Chang Y.J., Yu X.X., Xu L.P., Wang Y., Zhang X.H., Liu K.Y., Huang X.J. First-line therapy with donor-derived human cytomegalovirus (HCMV)-specific T cells reduces persistent HCMV infection by promoting antiviral immunity after allogenic stem cell transplantation. Clin. Infect. Dis. 2020;70:1429–1437. doi: 10.1093/cid/ciz368. [DOI] [PubMed] [Google Scholar]
- Zikos P., Van Lint M.T., Lamparelli T., Gualandi F., Occhini D., Mordini N., Berisso G., Bregante S., Bacigalupo A. A randomized trial of high dose polyvalent intravenous immunoglobulin (HDIgG) vs. cytomegalovirus (CMV) hyperimmune IgG in allogeneic hemopoietic stem cell transplants (HSCT) Haematologica. 1998;83:132–137. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original single-cell RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus repository under accession number: GSE192721.
This paper does not report the original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.