Abstract
Aroylated phenylenediamines (APDs) are novel modulators of innate immunity with respect to enhancing the expression of antimicrobial peptides and maintaining epithelial barrier integrity. Here, we present a new study on induction of autophagy in human lung epithelial cells by the APD HO53. Interestingly, HO53 affected autophagy in a dose-dependent manner, demonstrated by increased microtubule-associated proteins 1A/1B light-chain 3B (LC3B) processing in mature polarized bronchial epithelial cells. The quantification of LC3B puncta showed increased autophagy flux and formation of autophagosomes visualized by transmission electron microscopy. The phenotypic changes indicated that autophagy induction was associated with activation of 5′ adenosine monophosphate-activated protein kinase (AMPK), nuclear translocation of transcription factor EB (TFEB), and changes in expression of autophagy-related genes. The kinetics of the explored signaling pathways indicated on activation of AMPK followed by the nuclear translocation of TFEB. Moreover, our data suggest that HO53 modulates epigenetic changes related to induction of autophagy manifested by transcriptional regulation of histone-modifying enzymes. These changes were reflected by decreased ubiquitination of histone 2B at the lysine 120 residue that is associated with autophagy induction. Taken together, HO53 modulates autophagy, a part of the host defense system, through a complex mechanism involving several pathways and epigenetic events.
Keywords: Bronchial epithelium, Aroylated phenylenediamine, Adenosine monophosphate-activated protein kinase, Transcription factor EB, Epigenetics
Introduction
Autophagy is the essential adaptive process promoting cell survival and maintaining cell homeostasis in response to different stimuli from constantly changing environment. The prominent function of autophagy is to provide nutrients and energy for cellular processes during starvation. It is also cytoprotective in stress conditions such as hypoxia, shortage of growth factors, accumulation of misfolded proteins, and damaged organelles [1, 2]. Moreover, autophagy is considered as a selective process for innate and adaptive immunity, a part of cell autonomous defense system, protecting the host from pathogens [3, 4]. Interestingly, autophagy impairment is also related to autoimmune and inflammatory diseases such as inflammatory bowel diseases, e.g., Crohn's disease [5], implicating autophagy as a vital process with precise regulation [1, 4].
The epithelial layer in the human respiratory tract is the first line of defense, providing an important protective barrier for the host [6]. The initial active shield contains the mucus layer composed of different glycoproteins and antimicrobial peptides (AMPs) [6, 7] accompanied by mucociliary clearance [8]. However, when the first line of defense fails and bacteria enter epithelial cells, autophagy is a crucial process for protecting the host from invading pathogens [9, 10]. Dynamics of the autophagy process varies depending on the cell type. The clearance of pathogens is more rapid in phagocytic cells such as macrophages than in nonphagocytic bronchial epithelial cells [11]. Importantly, airway epithelial cells can effectively eliminate pathogens like Pseudomonas aeruginosa [12, 13]. Moreover, pathogens utilize a variety of virulence factors, allowing them to replicate within host cells, e.g., Mycobacterium tuberculosis (Mtb) blocks autophagosome maturation in infected macrophages [14]. Therefore, modulation of autophagy pathways can be an effective strategy to eliminate pathogens and to avoid/limit use of antibiotics, thereby reducing the selection of multidrug-resistant bacterial strains [15].
Several different agents including inducers of AMPs, phenylbutyrate, and the active form of vitamin D3 have been shown to induce autophagy and promote intracellular killing of Mtb in human macrophages [16, 17]. In addition, vitamin D3 and phenylbutyrate gave a positive outcome in the clinical trials for the treatment of pulmonary tuberculosis [18, 19]. Moreover, the short-chain fatty acid butyrate imprints an antimicrobial program in macrophages including activation of autophagy [20]. Recently, we reported on a compound from the novel group of aroylated phenylenediamines (APDs), HO53, that induced expression of several antimicrobial effectors, enhanced tight junctions and was efficient against P. aeruginosa infection [21].
In this study, we analyzed if HO53 induced autophagy in human airway epithelial cells and investigated molecular mechanisms behind autophagy induction by HO53. For this purpose, we utilized two human bronchial epithelial cell lines BCi-NS1.1 (hereafter BCi) and VA10. Both cell lines retain basal-like character and are able to differentiate toward polarized epithelium approaching mature epithelial cells [22, 23]. For differentiated BCi cells in air-liquid interphase (ALI) culture, autophagy induction with HO53 was monitored by analysis of microtubule-associated proteins 1A/1B light-chain 3B (LC3B) processing, the presence of LC3B puncta, and transmission electron microscopy (TEM). To define cell signaling pathway/s involved in induction of autophagy by HO53 in the ALI culture of BCi cells, we performed RNA sequencing (RNAseq) analysis. Further, we analyzed phosphorylation level of 5′ adenosine monophosphate-activated protein kinase (AMPK), nuclear translocation of the transcription factor EB (TFEB), and expression of autophagy-related genes. In addition, we evaluated if HO53 modulates autophagy through epigenetic events related to changes in gene expression of histone-modifying enzymes. We analyzed the expression of ubiquitin-specific peptidase 44 (USP44) and enhancer of zeste homolog 2 (EZH2) and if these transcriptional changes were reflected in ubiquitination status of histone 2B at lysine 120 residue (H2BK120) and trimethylation of histone H3 at lysine 27 residue (H3K27me3), respectively. Taken together, we evaluated induction of autophagy by HO53 in human airway epithelial cells and defined several signaling pathways and epigenetic events involved in this stimulation.
Materials and Methods
Reagents and Materials
Collagen from human placenta (C7521), the secondary antibody conjugated with horse radish peroxidase for Western blotting (A0545), DAPI (D9564), and all chemicals were purchased from Sigma. UltroserG (15950-017) was obtained from Pall Life Sciences and DMSO (sc-358801), Bafilomycin A1 (sc-201550), and rapamycin (sc-3504) from Santa Cruz. Secondary antibodies for immunofluorescence staining were obtained from Thermo Scientific (A-11070, A-11020, and A-21244). HO53 was synthesized as described previously [21].
Cell Cultures
The human bronchial epithelial cell line BCi-NS1.1 (BCi) immortalized with a retrovirus expressing human telomerase (hTERT) was from Dr Matthew S. Walters, Weill Cornell Medical College, New York, NY, USA [23]. An E6/E7 viral-oncogene-immortalized human bronchial epithelial cell line VA10 has been described previously [22]. Both cell lines were cultured in Bronchial/Tracheal Epithelial Cell Growth Medium (Cell Applications, 511A-500) supplemented with retinoic acid (Cell Applications, 511-RA) and penicillin-streptomycin (20 U/mL, 20 μg/mL, respectively; Life Technologies, 15140122) at 37°C and 5% CO2. The ALI culture of BCi and VA10 cells was maintained as described previously [22, 23]. Cells in monolayer were treated by direct addition of the compound to the culture medium. Differentiated cells were used for experiments after 21 days of the ALI culture, and the treatment compound was added to the lower chamber for indicated time.
Immunoblotting
Cells were washed with PBS and lysed in RIPA lysis buffer (Santa Cruz, sc-364162) supplemented with Halt Protease Inhibitor Cocktail (Thermo Scientific, 87786) and Phosphatase Inhibitor Cocktail (Cell Signal., 5870) on ice for 30 min. Histone extracts were obtained from ∼5 × 105 differentiated cells lysed in 80 μL of PBS with 0.5% Triton X-100 supplemented with protease and phosphatase inhibitors cocktail for 10 min on ice followed by overnight acid extraction (in 0.2 N HCl) of histones from cell pellets. Acid extracts containing histones were neutralized by addition of 2 M NaOH (1/10 of final volume). The total protein of 10–20 μg and 2.5 μg of purified histone extracts was separated using NuPAGE 4–12% Bis-Tris gradient gels (Life Technologies, NP0323) and NuPAGE MES SDS Running Buffer (Life Technologies, NP0002) or polyacrylamide gel electrophoresis and SDS-Tris-glycine buffer (25 mM Tris, 250 mM glycine, and 0.5% SDS) with the running conditions 120 V and 275 mA. As described previously [21], the proteins were transferred on to a polyvinylidene fluoride membrane (0.2 μm pores) using XCell IITM Blot Module (Invitrogen, EI9051), and the membrane was blocked with 10% skimmed milk or 10% BSA in TBS-T buffer (50 mM Tris, 150 mM NaCl, 0.1% Tween-20) for 1 h at room temperature. Then, the membrane was incubated with primary antibodies (shown in online suppl. Table S1; for all online suppl. material, see www.karger.com/doi/10.1159/000521602) overnight at 4°C using a dilution recommended by the manufacturer's protocol. After washing with TBS-T buffer, the membrane was incubated with horse radish peroxidase-conjugated secondary antibodies (1:10,000 dilution) in 5% skimmed milk or 5% BSA in TBS-T for at least 1.5 h at room temperature. Immunoblots were developed using Pierce ECL Plus Western Blotting Substrate (Thermo Scientific, #34095) or Western Blotting Luminol Reagent (Santa Cruz, sc-2048) and the ImageQuant LAS 4000 system (GE Healthcare). Quantification of the band intensity was performed using ImageJ software.
Immunofluorescence
Cells growing on ALI filters were fixed using prechilled methanol at 4°C overnight and then by prechilled acetone. Fixed cells on ALI filters were hydrated in IF buffer (0.3% Triton X-100 in PBS) and incubated with the blocking buffer (10% FBS in IF buffer) and then with primary antibodies (shown in online suppl. Table S1) overnight at 4°C using concentration recommended by the manufacturer's protocol. The next day, filters were washed and incubated with DAPI (1:5,000) and secondary antibodies Alexa Fluor 488 (A-11070) and/or Alexa Fluor 594 (A-11020) for 2 h. Cells growing on ALI filters were mounted in FluoromountTM Aqueous Mounting Medium (F4680 Sigma), and coverslips were placed over the filters. Images were taken using Olympus fluoview FV1200 confocal microscope at 30× and 60× magnification followed by ImageJ analysis. The randomized blind counting of LC3B puncta was performed manually from at least 3 different random areas of each sample. Nuclear translocation of TFEB and co-localization of LC3B with mucous and ciliated cells were evaluated by using the “Colocalisation” function of the Olympus fluoview FV1200 confocal microscope software. The threshold was adjusted to value of 1,250, showing signal only from co-localized TFEB/nuclei (pink) extracted from merged images followed by quantification of pixel intensity by ImageJ software. The same parameters of image acquisition and threshold were used for all images.
Transmission Electron Microscopy
BCi-NS1.1 cells growing on trans-well inserts in ALI conditions were fixed with 2.5% glutaraldehyde (Ted Pella, Inc.) by addition of fixative to the upper and lower chamber. Next, cells were washed in phosphate buffer (0.075 M with 0.15 M sucrose) twice for 2 min and postfixed in 2% osmium tetroxide (J.B. EM Services Inc.) for 30 min followed by washing twice for 3 min. Cells were dehydrated in increasing concentrations of ethanol: 25%, 50%, and 70% for 2 min each. Then, alcohol was replaced with 4% uranyl acetate in 70% ethanol (J.B. EM Services Inc.) for 7 min, followed by 2 min incubations in 80%, 90%, and 96% solutions of ethanol and at the end, in pure ethanol for 5 min and twice for 7 min. Then, the trans-well filters were placed on the coverslips with a drop of resin (Spurr Resin; Ted Pella, Inc.), and cells were embedded by adding a few drops of resin on top of the cell layer and incubated for 2 h at room temperature. Gelatin capsules were filled with resin and placed upside down on top of the coverslips to create a block and incubated overnight at 70°C. Afterward, resin blocks and filters were separated from coverslips by submerging them for few seconds in liquid nitrogen. After that, plastic filters were broken away which created round-shaped resin disks. Ultrathin (100 nm) sections were cut with diamond knife (45° DiATOME) on an ultramicrotome (Leica EM UC7) and placed on copper grids (Ted Pella, Inc.). Sections on grids were stained for 1 min with lead citrate (3% Ultrostain 2, Leica) and imaged using a JEM-1400PLUS PL Transmission Electron Microscope at various magnifications.
Cytotoxicity Assay
The cytotoxic effect of Bafilomycin A1 on bronchial epithelial BCi cells differentiated in an ALI culture was assessed by measurements of lactic dehydrogenase release from the cells in both the apical (upper, with cells) and basolateral (lower) side of the ALI filters. Bafilomycin A1 at 100 nM was added to the lower chamber of the trans-well insert (basolateral side), while standard cell culture medium was applied on the apical side for 24 h. Cytotoxicity of bafilomycin was measured using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega) with modified incubation time of 15 min. Cytotoxicity was calculated as a percentage of lactic dehydrogenase release in the positive control (100%).
RNA Analysis
Total RNA was extracted using the NucleoSpin RNA kit (MACHEREY-NAGEL, 740955.50). Total RNA of 1 μg was used to synthesize complementary DNA (cDNA) according to the manufacture's recommendations using the High-Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems, 4368814). One µl of cDNA, 5 μL of PowerUp SYBR Green Master Mix (Applied Biosystems, A25742), and 0.5 μM primers (shown in online suppl. Table S2) were used for qRT-PCRs. qRT-PCRs were performed using the LG 7500 Real-Time PCR System (Applied Biosystems) with the following cycling conditions: (1) holding stage: 95°C for 10 min, followed by 40 cycles of (2) denatured stage: 95°C for 15 s and (3) annealed/extended stage: 60°C for 1 min. The 2(−ΔΔCT) Livak method was utilized for calculating fold differences over untreated control [24].
Preparation of Poly-A cDNA Sequencing Libraries
As described previously [25, 26], the quality (RIN score) and quantity of isolated total RNA samples were assessed using the DNA 5K/RNA chip for the LabChip GX (PerkinElmer). cDNA libraries derived from poly-A mRNA were generated using Illumina's TruSeq RNA v2 Sample Prep Kit. Briefly, poly-A mRNA was isolated from total RNA samples (0.2–1 μg input) using hybridization to poly-T beads. The poly-A mRNA was fragmented at 94°C, and first-strand cDNA was prepared using random hexamers and the SuperScript II Reverse Transcriptase (Invitrogen). Following second-strand cDNA synthesis, end repair, addition of a single A base, indexed adapter ligation, AMPure bead purification, and PCR amplification, the resulting cDNA sequencing libraries were measured on the LabChip GX, diluted to 3 nM and stored at −20°C.
RNAseq and Gene Expression Analysis
Samples were pooled and sequenced on NovaSeq 6000 (24 samples/pool/lane) using on-board clustering. Paired-end sequencing (2 × 125 cycles) was performed using S4 flowcells, following the XP workflow. We quantified the RNA transcript expression with Kallisto (version 0.45.0) [27] using the Homo sapiens GRCh38 reference transcriptome [28]. Gene expression estimates were computed with the sleuth R package (v0.30) [29]. Gene set enrichment analysis (GSEA; version 3.0) developed by Broad Institute [30] was used for gene ontology analysis. The differentially expressed genes were ranked on the list based on the expression (b-estimate) and q value. Next, the ranked lists of differentially expressed genes were analyzed by GSEA software using the “hallmark gene sets” database of biological processes and the customized list of autophagy-lysosome genes [31] as a reference with the following parameters: (1) number of permutations: 1,000, (2) enrichment statistic: classic, and (3) gene sets larger than 5,000 and smaller than 15 were excluded from the analysis. Among positively and negatively enriched gene sets, autophagy-related pathways with a false discovery rate <0.05 were presented. Heatmaps showing significant (p < 0.05) gene expression presented as log2 fold change were created in R studio software.
Statistical Analysis
Results are presented as mean ± standard error of mean from at least three independent experiments. One- and two-way ANOVA and the student t-test were used to determine significance of the data; p values and corrections for multiple comparisons are indicated in the figure legends. The statistical analysis was performed with GraphPad Prism 6 software (GraphPad, San Diego, CA, USA). Western blots are representative of at least three independent experiments, except online supplementary Figure S4A.
Results
HO53 Induces Autophagy in Mature Human Lung Epithelial Cells
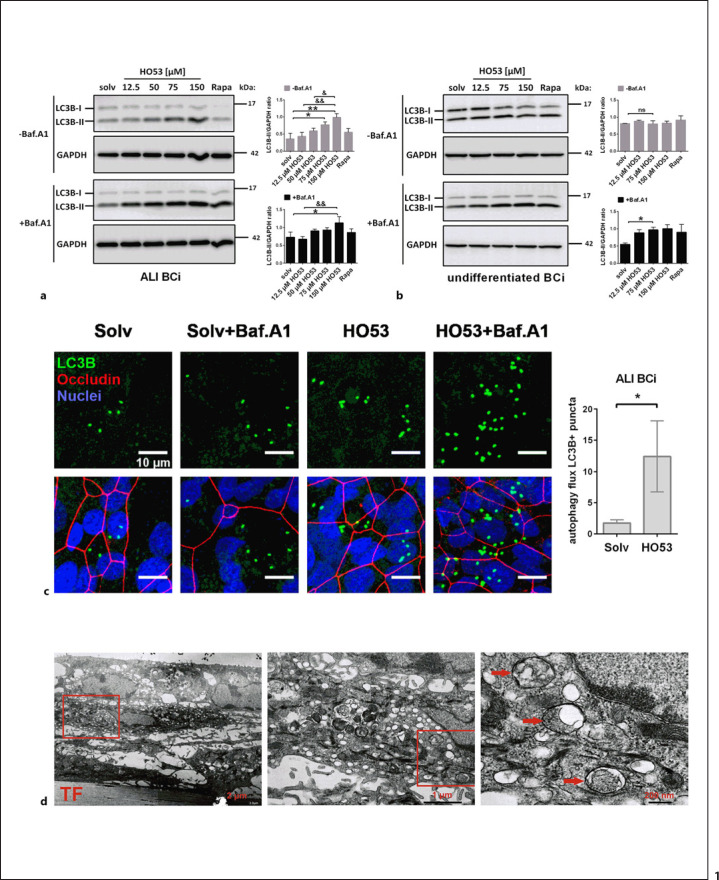
Several markers are utilized to monitor autophagy induction. The most common is the autophagosomal marker LC3B, more precisely, processing of the soluble LC3B-I to autophagosome-associated LC3B-II, indicating the formation of autophagosomes. Further, degradation of the autophagy-substrate p62, as well as Bafilomycin A1, an inhibitor of lysosomal acidification and autophagosome-lysosome fusion, is used to evaluate lysosomal degradation of autophagosomal cargo described as the autophagy flux [32]. To assess the effect of the APD compound HO53 on autophagy induction in the human bronchial epithelial cell lines BCi and VA10, we analyzed LC3B processing and visualized the presence of LC3B puncta and the occurrence of autophagosomes using TEM. Both human bronchial epithelial cell lines can be used as undifferentiated monolayer cells or mature differentiated cells in ALI cultures, forming polarized epithelial layers [22, 23]. First, we analyzed the effect of HO53 on autophagy induction by analysis of LC3B processing in both ALI and undifferentiated cells (shown in Fig. 1a, b). Treatment with increasing doses of HO53 for 24 h led to dose-dependent accumulation of LC3B-II in ALI BCi (shown in Fig. 1a) but not in undifferentiated cells (shown in Fig. 1b). Further, in co-treatment with Bafilomycin A1 applied for 24 h, without presenting any significant cytotoxic effect on the cells (shown in online suppl. Fig. S2), these changes were significant also in undifferentiated cells for 75 μM HO53 (shown in Fig. 1b) but in ALI cells only, at 150 μM (shown in Fig. 1a). These results indicate that autophagy is induced in the mature polarized BCi cells. A similar phenotype of autophagy induction by HO53 linked to cell differentiation status was observed in VA10 cells (shown in online suppl. Fig. S1A, S1B), where autophagy induction was identified only in mature cells (shown in online suppl. Fig. S1A). We observed a low expression level of p62 protein, an additional autophagy flux indicator in both human lung epithelial cell lines VA10 and BCi, and the expression remained unchanged upon treatment with HO53 (shown in online suppl. Fig. S1A–D, respectively). Considering the pronounced effect of HO53 on the conversion of LC3B-I to the lipidated LC3B-II form in BCi ALI cells, we continued our studies with ALI cells resembling in vivo environment. Based on the processing of LC3B-I to LC3B-II and our previous studies [21], we selected 75-μM HO53 concentration for further studies. Fluorescent LC3B puncta are commonly used as an indicator for autophagosomes [32]. The treatment with HO53 increased the number of LC3B puncta in ALI cells co-stained with occludin, a tightjunction protein serving as a marker for differentiated cells (shown in Fig. 1c). The quantification of LC3B puncta (LC3B+) in cells with and without Bafilomycin A1 indicated that the treatment with HO53 induced autophagy flux in differentiated BCi cells (shown in Fig. 1c). Interestingly, the LC3B puncta were mainly clustered in small groups, suggesting that autophagy possibly occurs in specific cell types of ALI differentiated cell layers. Therefore, we next co-stained LC3B with mucin 5AC (MUC5AC), a marker for mucous cells, and acetylated (at K40) tubulin α A4 (TUBA4A acetyl K40), a marker for ciliated cells, both occurring in ALI-differentiated cell layers (shown in online suppl. Fig. S1E, S1F). We observed co-localization of LC3B and MUC5AC in specific areas but occurrence of LC3B puncta clusters was not exclusively restricted to mucous cells (shown in online suppl. Fig. S1E). Next, TEM analysis of ALI cells treated with HO53 and Bafilomycin A1 revealed the presence of typical double-membrane-limited autophagosomes (shown in Fig. 1d) characteristic for autophagy induction. Taken together, our findings show that HO53, an innate immunity modulator, stimulates autophagy in human lung epithelial cells.
Fig. 1.
HO53 treatment induces autophagy in human airway epithelial cells. BCi cells were stimulated for 24 h with different doses of HO53, and 250 nM Rapa was used as a positive control for autophagy induction, and DMSO (final concentration of 0.3%) was used as a solv, all in combination with (+Baf.A1) or without (−Baf.A1) Bafilomycin A1 (100 nM). Treatment of differentiated BCi cells was performed by addition of the compound to the lower chamber of the trans-well insert. Induction of autophagy in the ALI culture (a) and undifferentiated BCi (b) was evaluated by analysis of LC3B processing on Western blotting. The processing of LC3B-I to LC3B-II was quantified by measurement of the LC3B-II band intensity versus GAPDH loading control and presented as the LC3B-II/GAPDH ratio. Data present average ± SEM from n = 3 independent experiments analyzed by one-way ANOVA with Sidak post hoc test, where *p < 0.05, **p < 0.01 and ns versus solvent control, &p < 0.05 versus 50 μM HO53, &&p < 0.01 versus 12.5 μM HO53. Samples were run in one experiment on separate gels/blots processed in parallel, and full-length blots are presented in Supplementary Figure S7. cAnalysis of the autophagy induction by HO53 (75 μM) in ALI-cultured BCi cells by immunostaining of LC3B puncta (green), nuclei (blue), and occludin (red), a tight-junction protein characteristic for the differentiated BCi cells in the ALI culture. The scale bar is 10 μm. Autophagy flux in ALI-cultured BCi was calculated based on number of LC3B+ (positive) puncta using the formula: (sample + Baf.A1)/sample presented as autophagy flux LC3B + puncta. Data present average ± SEM from n = 5 independent experiments analyzed by an unpaired t-test, where *p < 0.05. dTEM analysis of differentiated BCi cells treated with HO53 (75 μM) and Bafilomycin A1 (100 nm) for 24 h. Autophagosomes are indicated by red arrows. TF indicates the trans-well filter/insert; red squares indicate a magnified area with scale bars for images as indicated (from left to right) 2 μm, 1 μm and 200 nm. Representative images of n = 4 trans-well inserts from 2 independent experiments. ns, nonsignificant; solv, solvent control; Rapa, rapamycin; SEM, standard error of mean.
HO53 Treatment Alters Autophagy-Related Pathways in Mature Human Bronchial Epithelial Cells (BCi)
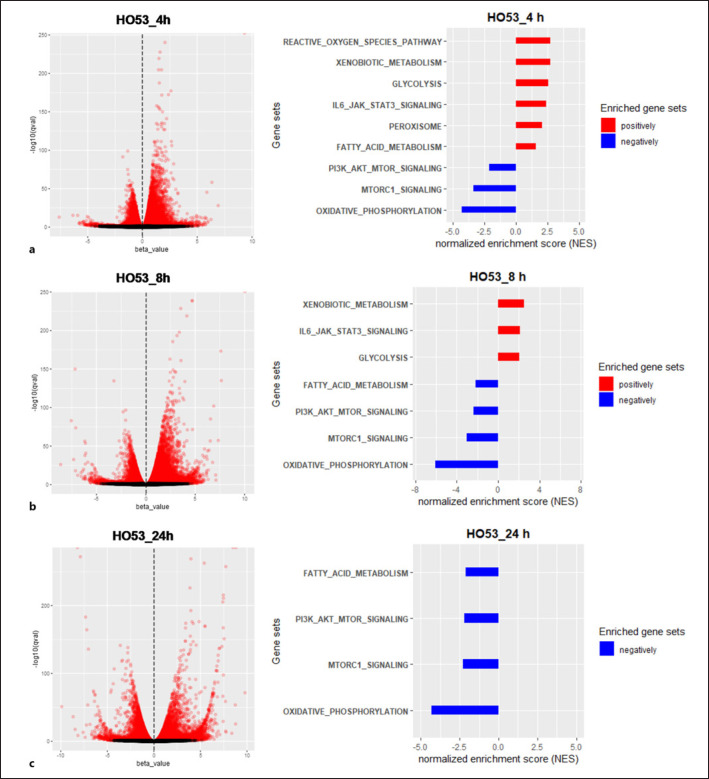
To resolve the mechanism behind induction of autophagy by HO53 in ALI BCi cells, we performed RNAseq analysis of the transcriptome at different time points. Based on our previous studies, where we observed HO53 affecting innate immunity and epithelial barrier integrity [21], we selected 4 h, 8 h, and 24 h time points of HO53 treatment. Analysis of the transcriptome of ALI BCi cells treated with HO53 revealed a broad response at the RNA level that was observed after 4 h treatment and expanded with time (shown in Fig. 2, online suppl. S3A). GSEA revealed that several gene sets were affected by HO53 (shown in online suppl. Fig. S3B) and among them were pathways related to autophagy [33, 34, 35, 36, 37, 38, 39, 40] (shown in Fig. 2). After 4 h treatment with HO53, we observed molecular signature for the following autophagy-related pathways that were positively correlated to the gene set enrichment (upregulated): (1) reactive oxygen species (ROS) [36], (2) xenobiotic metabolism [38], (3) glycolysis [34], (4) interleukin 6-Janus kinase-signal transducer and activator of transcription 3 (IL6-JAK-STAT3) [37], (5) peroxisome [33, 39], and (6) fatty-acid metabolism [33, 39]. Moreover, the negatively correlated pathways (downregulated) were the following: (7) phosphoinositide 3-kinase-protein kinase B-mechanistic target of rapamycin kinase (PI3K-AKT-mTOR) [40], (8) mechanistic target of rapamycin kinase complex 1 (mTORC1) [40], and (9) oxidative phosphorylation (OXPHOS) [35, 36] (shown in Fig. 2a). With time, the molecular signatures were altered, and ROS and peroxisome pathways were not enriched in the GSEA categories at 8 and 24 h (shown in Fig. 2b, c). Furthermore, the pathway linked to fatty-acid metabolism was negatively correlated after 8 h (shown in Fig. 2b). We observed only four negatively correlated pathways linked to mTOR/mTORC1 signaling, fatty-acid metabolism, and OXPHOS after 24 h (shown in Fig. 2c). Importantly, mTOR/mTORC1 signaling and OXPHOS pathways were negatively enriched for all time points. Overall, a broad effect of HO53 on the gene expression in human lung epithelial BCi cells shows molecular signature for pathways involved in autophagy regulation.
Fig. 2.
HO53 affects autophagy-related pathways in mature human bronchial epithelial cells (BCi). Volcano plots with differentially expressed transcripts and GSEA of the expression data from BCi cells differentiated in an ALI culture after 4 h (a), 8 h (b), and 24 h of treatment with HO53 (75 μM) (c). Volcano plots represent b-estimate (expression) versus log of significance (−log10[q]) from the Kallisto/sleuth Wald test. Significant differentially expressed genes were marked in red based on q ≤ 0.05. In pathway analysis (GSEA), autophagy-related gene sets were presented as positively correlated gene sets (red, NES >0) and negatively correlated gene sets (blue, NES <0) with FDR ≤0.05. FDR. FDR, false discovery rate; NES, normalized enrichment score.
HO53 Activates the AMPK Pathway Linked to Autophagy
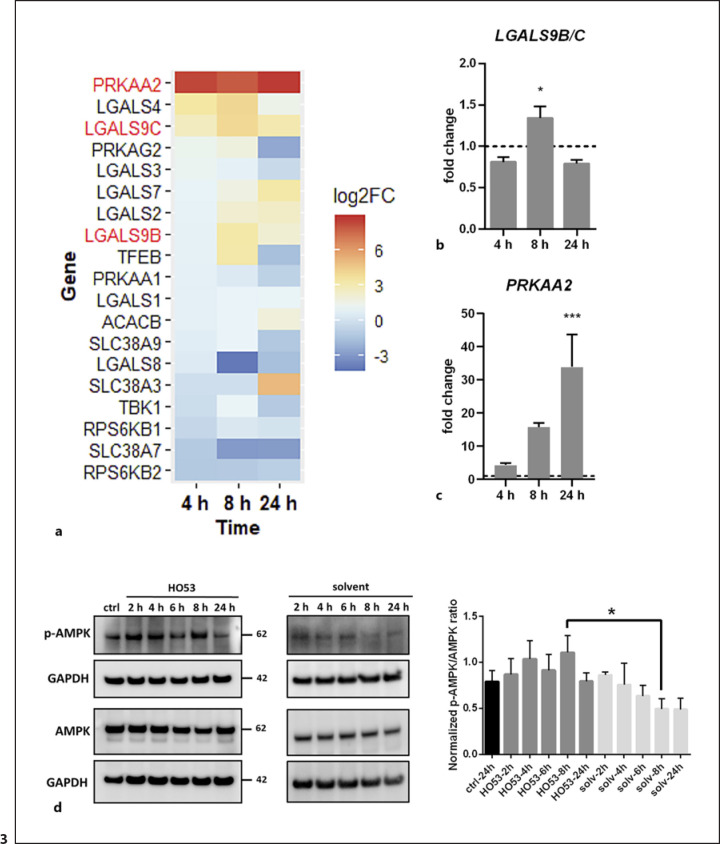
In our previous studies, we demonstrated that HO53 increases phosphorylation of STAT3 in the monolayer BCi, and the STAT3 pathway has been shown to be involved in the autophagy process [21]. Therefore, we first analyzed if HO53 has the same effect on posttranslational modifications of STAT3 in ALI BCi cells and if these changes contribute to autophagy induction. Our results suggest that treatment with HO53 did not affect posttranslational modifications of STAT3 related to autophagy induction in differentiated BCi cells (shown in online suppl. Fig. S4A). Galectins have been shown to act as a cytoplasmic sensor of intracellular pathogens, leading to activation of autophagy [41]. In addition, galectins can control mTOR and AMPK to induce autophagy upon endolysosomal membrane damage [42, 43]. Interestingly, analyses of the RNAseq data revealed enhanced expression of genes encoding galectins, e.g., LGALS4, LGALS9B/C, LGALS7 (shown in Fig. 3a), and other genes encoding key proteins involved in the mechanism of autophagy induction initiated by galectins [42] (shown in Fig. 3a). Notably, the expression pattern of LGALS9B/C encoding galectin 9 observed in RNAseq data was confirmed by qPCR, demonstrating significant increase after 8 h (shown in Fig. 3b). However, these changes were not reflected by galectin 9 at the protein level as we detected a slight increase of galectin 9 at later time points (shown in online suppl. Fig. S4B). Furthermore, we observed remarkably high expression of PRKAA2 encoding catalytic subunit α2 of AMPK for all time points of HO53 treatment (shown in Fig. 3a). We confirmed this observation by qPCR, noting 4-, 15-, and 33-fold increase in PRKAA2 gene expression at 4 h, 8 h, and 24 h, respectively (shown in Fig. 3c). To test whether these transcriptional changes caused by HO53 contribute to the activation of AMPK and inhibition of mTOR/mTORC1 indicated in the pathway analysis, we examined phosphorylation status of AMPK (Thr172) (shown in Fig. 3d) and the downstream target of mTOR and ribosomal protein S6 kinase B1 (S6K1) (Thr389) (shown in online suppl. Fig. S4C). We measured p-AMPK/AMPK and p-S6K1/S6K1 ratios at 2, 4, 6, 8, and 24 h of treatment with HO53 and found an increased phosphorylation level of AMPK at 8 h (shown in Fig. 3d). We also observed a decreased but not significant phosphorylation level of S6K1 at 4 h (shown in online suppl. Fig. S4C). Together, these results suggest that HO53 induces autophagy through a complex mechanism involving many cellular pathways linked to AMPK and possibly mTOR signaling.
Fig. 3.
HO53 activates the AMPK signaling cascade in differentiated BCi cells. Differentiated BCi cells in the ALI culture were treated with HO53 (75 μM) for 4 h, 8 h, and 24 h. aHeatmap showing changes in expression as log2FC of genes related to the mTOR/AMPK pathway at 4 h, 8 h, and 24 h of treatment with HO53. The heatmap shows significantly differentially expressed genes in comparison to control cells collected at the same time points (p < 0.05). Genes selected for further analysis were marked in red. Nonsignificant differentially expressed genes have arbitrary value 0. Expression level of LGALS9B/C (b) and PRKAA2 (c) genes analyzed by qRT-PCR. Gene expression was represented as a fold change in comparison to control cells (value 1, dashed line) collected at the same time point as treated cells and normalized to the EEF2 reference gene. n = 3 trans-well inserts from 3 independent experiments ± SEM. *p < 0.05, ***p < 0.001 versus control cells analyzed by one-way ANOVA with the Dunnett post hoc test. dActivation of the AMPK signaling pathway analyzed by Western blotting of phosphorylated AMPKα (Thr172) at different time points of HO53 treatment. Quantification of Western blots was performed in two steps: first, by measurements of band intensity and normalization to GAPDH (loading control) and then by calculation of the normalized ratio for phospho-AMPKα (Thr172) to total AMPKα. The representative Western blot of n = 4 independent experiments. Quantitative comparison was done on the samples run in one experiment on separate gels/blots processed in parallel. Statistical analysis was performed by one-way ANOVA with the Sidak post hoc test, where *p < 0.05 versus corresponding solvent control. Full-length blots are presented in online supplementary Figure S7. log2FC, log2 fold change; SEM, standard error of mean.
HO53 Treatment Leads to TFEB Nuclear Translocation and Augments Expression of Autophagy-Associated Genes
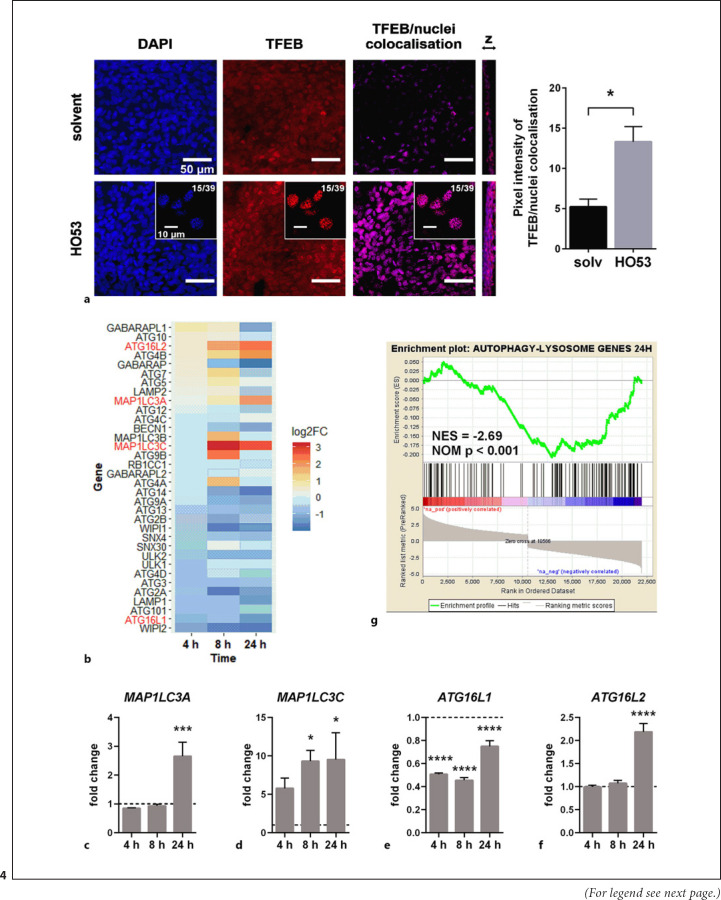
We next monitored TFEB nuclear translocation responsible for transcriptional regulation of autophagy and lysosomal gene expression. Moreover, RNAseq analysis indicated that HO53 affected the mTOR pathway, well known as a negative regulator of TFEB translocation [44, 45]. We observed increased nuclear localization of TFEB at 24 h of treatment with HO53 (shown in Fig. 4a). Only weak nuclear signal for TFEB was detected at earlier time points of 4 and 8 h (shown in online suppl. Fig. S5A, S5B), suggesting that TFEB nuclear translocation is a late event of the HO53-induced signaling cascade, leading to autophagy induction. We also assessed whether HO53 treatment of mature BCi cells led to the differential expression of autophagy-related genes. By using the list of autophagy-related genes (HUGO Gene Nomenclature Committee [HGNC] Group ID 1022) [46] from HGNC database [47, 48] as a reference, we analyzed their expression in the RNAseq data (shown in Fig. 4b) and confirmed expression of selected genes by qPCR (shown in Fig. 4c-f). Interestingly, the HO53 compound enhanced expression of the genes encoding LC3 isoforms, especially MAP1LC3A encoding LC3A at 24 h (shown in Fig. 4c) and MAP1LC3C encoding LC3C at 8 h and 24 h (shown in Fig. 4d). Further, HO53 treatment affected expression of ATG16L genes in differentiated BCi cells, encoding a component of E3-like complex that couples ATG8 family proteins to phosphatidylethanolamine [1]. Interestingly, the expression of ATG16L1 decreased with time, but the expression of its homolog ATG16L2 was increased in analyzed RNAseq data (shown in Fig. 4b). The same expression pattern for ATG16L1 and ATG16L2 was reconfirmed by qPCR, where expression of ATG16L1 was downregulated during the HO53 treatment (shown in Fig. 4e), and ATG16L2 was upregulated after 24 h (shown in Fig. 4f). These results indicate that HO53 affects nuclear translocation of TFEB and induces expression of selected autophagy-related genes associated with formation of autophagosomes at the early phase of the autophagy process. Notably, several autophagy-related genes were downregulated (shown in Fig. 4b). To evaluate if expression of lysosomal genes was affected by HO53, we performed gene set enrichment analysis for the set of autophagy-lysosomal genes at 24 h (shown in Fig. 4g) [31]. The enrichment plot for autophagy-lysosomal genes was negatively correlated, indicating that other mechanisms partially interacted with the effect of TFEB.
Fig. 4.
HO53 treatment leads to nuclear translocation of TFEB and augments expression of autophagy-related genes in differentiated BCi cells. aAnalysis of TFEB nuclear translocation upon treatment with 75 μM HO53 after 24 h by confocal imaging of nuclei (blue, DAPI), TFEB (red), and extracted signal from co-localized TFEB/nuclei (pink) from merged channels with scale bar 50 μm. Representative single slices from z-stacked images (15/39) were placed for each channel of HO53-treated samples with scale bar 10 μm. The representative images from at least 3 different areas of the ALI filter from n = 3 independent experiments with quantification of pixel intensity for a signal from co-localized TFEB/nuclei (pink) extracted from merged DAPI/TFEB images ± SEM. Statistical analysis was performed by using an unpaired t-test with Welch's correction, *p < 0.05. Changes in autophagy-related genes expression in differentiated BCi cells in ALI at 4 h, 8 h, and 24 h of treatment with HO53. bA heatmap showing the expression level represented as a log2FC of significantly differentially expressed genes in comparison to control cells collected at the same time points (p < 0.05). Genes selected for further analysis were marked in red. Nonsignificant differentially expressed genes have arbitrary value 0. Expression level of selected autophagy-related genes: MAP1LC3A (c), MAP1LC3C (d), ATG16L1 (e), and ATG16L2 (f) analyzed by qRT-PCR. Gene expression was represented as a fold change in comparison to control cells (value 1, dashed line) collected at the same time point as treated cells and normalized to the EEF2 reference gene. n = 3 trans-well inserts from 3 independent experiments ± SEM. *p < 0.05, ***p < 0.001, ****p < 0.0001 versus control cells analyzed by one-way ANOVA with the Dunnett post hoc test. gEnrichment plot for autophagy-lysosomal genes obtained from GSEA of the expression data from differentiated BCi cells in the ALI culture after 24 h of treatment with HO53. NES and NOM p are indicated on the plot. log2FC, log2 fold change; NES, normalized enrichment score; NOM p, nominal p value; SEM, standard error of mean.
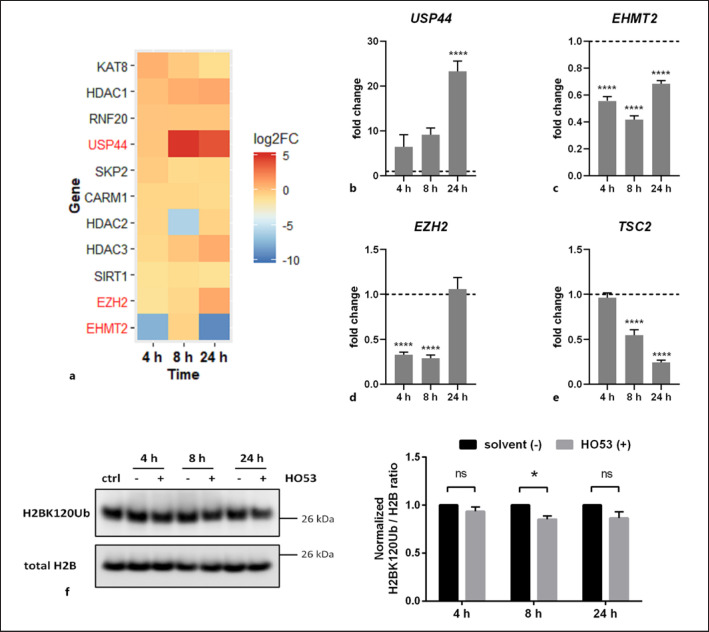
HO53 Modulates Epigenetic Changes Involved in Autophagy Induction
Treatment with HO53 induces a broad response in gene expression and affects several autophagy-related pathways in differentiated human lung epithelial cells. Therefore, we hypothesized that HO53 modulates epigenetic events such as histone modifications, leading to autophagy induction. It has been shown that epigenetic enzymes like histone deacetylases, methyltransferases, and ubiquitinases can affect regulatory pathways for autophagy [49, 50]. We analyzed if HO53 can modulate expression of histone-modifying enzymes. Interestingly, USP44 expression encoding USP44, a key enzyme contributing to enhanced autophagy by deubiquitination of H2BK120Ub [51], was strongly upregulated after 8 h and 24 h of HO53 treatment (shown in Fig. 5a). The enhanced expression of USP44 was reconfirmed by qPCR (shown in Fig. 5b). Moreover, expression of EHMT2 encoding euchromatic histone lysine methyltransferase 2 (G9a), responsible for dimethylation of lysine 9 residue on histone 3 (H3K9me2), resulting in inhibition of autophagy [52], was downregulated to a different extent at all analyzed time points of HO53 treatment (shown in Fig. 5a). This was in agreement with expression of EHMT2 analyzed by qPCR (shown in Fig. 5c) and could promote autophagy by suppressing the inhibitory effect. Another enzyme repressing autophagy by trimethylation of H3K27me3 is EZH2 [50]. Based on RNAseq data, the expression of EZH2 was downregulated at the beginning of HO53 treatment and then enhanced after 24 h (shown in Fig. 5a). This suppression of EZH2 expression was confirmed at 4 h and 8 h by qPCR and reached a similar level as in untreated BCi cells after 24 h (shown in Fig. 5d). The kinetics of transcriptional changes for EZH2 was reflected in expression of TSC2 encoding tuberin, a downstream target gene of EZH2 (shown in Fig. 5e). Expression of TSC2, a negative regulator of mTOR, is regulated by EZH2 that binds to the TSC2 promoter and silences TSC2 transcription, leading to activation of the mTOR pathway and inhibition of autophagy [50]. Unlike EZH2, expression of TSC2 was gradually decreased with time of HO53 treatment (shown in Fig. 5e). In the continuation, we verified if histones modification status reflects transcriptional changes in expression of key epigenetic enzymes regulating autophagy. Consequently, we analyzed ubiquitination status of H2BK120 and trimethylation of H3K27 over time of HO53 treatment (shown in Fig. 5f, online suppl. S6). Interestingly, in bronchial epithelial cells, we observed a reduced level of ubiquitinated H2BK120 at 8 h as a result of HO53 treatment, indicating autophagy induction (shown in Fig. 5f). The level of trimethylated H3K27 in BCi cells remained unchanged upon treatment with HO53 at all analyzed time points (shown in online suppl. Fig. S6). In summary, these results indicate that HO53 affects expression of histone-modifying enzymes, precisely controlling epigenetic machinery responsible for the balance between induction and inhibition of autophagy.
Fig. 5.
HO53 treatment modulates epigenetic regulation of autophagy in ALI-cultured BCi cells. Differentiated BCi cells in the ALI culture were treated with HO53 (75 μM) for 4 h, 8 h, and 24 h. aHeatmap showing changes in expression of genes encoding epigenetic enzymes involved in regulation of autophagy presented as log2FC at 4 h, 8 h, and 24 h of treatment with HO53. The heatmap shows significant differentially expressed genes in comparison to control cells collected at the same time points (p < 0.05). Genes selected for further analysis were marked in red. Nonsignificant differentially expressed genes have arbitrary value 0. Expression level analyzed by qRT-PCR for the genes USP44 (b), EHMT2 (c), EZH2 (d), and TSC2 (e), a downstream target of the EZH2 protein. Gene expression was represented as a fold change in comparison to control cells (value 1, dashed line) collected at the same time points as treated cells and normalized to the EEF2 reference gene. n = 3 trans-well inserts from 3 independent experiments ± SEM ****p < 0.0001 versus control cells analyzed by one-way ANOVA with the Dunnett post hoc test. fA representative Western blot of H2BK120 ubiquitination and the total H2B level (loading control) analyzed by Western blot analysis in the ctrl, treated with solvent (−) and 75 μM HO53 (+) BCi cells in ALI at indicated time points. DMSO at final concentration of 0.3% was used as a solvent control. The Western blot band intensity was quantified as the ratio of H2BK120Ub to total H2B and normalized to solvent control (value 1) based on n = 3 independent experiments ± SEM. Quantitative comparison was done on the samples run in one experiment on separate gels/blots processed in parallel. Statistical analysis was performed by two-way ANOVA with Bonferroni's multiple comparisons test, *p < 0.05. Full-length blots are presented in online supplementary Figure S7. log2FC, log2 fold change; ctrl, control; SEM, standard error of mean.
Discussion
Autophagy as an integral part of innate immunity is recognized as one of the main host defense systems controlling invading pathogens (xenophagy). In response to the host defense mechanisms, several pathogens have evolved a variety of different strategies to escape from autophagy clearance. Therefore, development of novel autophagy activating molecules for the enhancement of innate immunity defenses against invading pathogens would be a beneficial therapeutic approach to treat infections.
Here, we present a novel innate immunity inducer, HO53, from the class of APD compounds in relation to autophagy in human airway epithelial cells. Treatment with HO53 of mature bronchial epithelial cells cultured in an ALI model, resembling the in vivo situation, led to increased LC3B-II accumulation together with the presence of LC3B puncta and autophagosomes visualized by TEM analysis. Based on these, we conclude that HO53 stimulates autophagy by promoting autophagosome formation in mature bronchial epithelial cells. Analysis of autophagy flux by measuring levels of p62 to evaluate lysosomal degradation of autophagosomal cargo revealed that levels of p62 in both undifferentiated and ALI cells remained unchanged. Thus, we used an additional approach, the co-treatment with Bafilomycin A1, an inhibitor of autophagosome-lysosome fusion. The quantification of LC3B puncta with and without Bafilomycin A1 showed increased autophagy flux in cells stimulated with HO53. The effect of HO53 on autophagy initiation in the ALI model seems to be similar to the mechanism of AMPK, promoting autophagy by priming autophagy kinases upon detection of bacterial outer-membrane vesicles independently on bacterial invasion [53]. Importantly, we showed previously that HO53 can induce production of antimicrobial effectors by the innate immunity system and enhance epithelial barrier integrity [21]. This, together with induction of autophagy could provide prevention and/or treatment of infectious diseases, especially caused by pathogens affecting initiation of autophagosome formation.
Furthermore, to exploit molecular mechanisms behind the observed induction of autophagy, we performed RNAseq analysis of mature bronchial epithelial cells treated with HO53 for different time points. The early response to HO53 resulted in enrichment of the following pathways directly related to autophagy: ROS, xenobiotic metabolism, glycolysis, IL6-JAK-STAT3, peroxisome, fatty-acid metabolism, PI3K-AKT-mTOR, and OXPHOS. Shen et al. [54] showed that phosphorylation of STAT3 at Y705 is considered as a stimulus for autophagy induction. Phosphorylated STAT3 was not able to interact with the catalytic domain of the eukaryotic translation initiation factor 2 α kinase 2 that consequently allowed for phosphorylation of eukaryotic translation initiation factor 2A and activation of autophagy [37, 54]. In our study, we observed enrichment in the IL6-JAK-STAT3 pathway at early time points of HO53 treatment. However, we did not observe any changes in phosphorylation of STAT3 at Y705 in mature BCi cells, suggesting that the STAT3 pathway is not involved in the autophagy induction in mature bronchial epithelial cells. Moreover, our results suggest that HO53 affects phosphorylation of STAT3 depending on the cell differentiation status because unlike in monolayer BCi cells used in our previous work [21], we did not observe any changes in mature BCi cells. Furthermore, stimulation of autophagy by pro-inflammatory stimuli is unlikely because our previous work showed that unlike in monolayer BCi cells, HO53 did not induce pro-inflammatory cytokines/chemokines in ALI-cultured cells at the protein level [21].
Moreover, HO53 treatment of mature BCi cells affected the ROS pathway that could contribute to further induction of autophagy because ROS are known autophagy mediators in response to oxidative stress or in infection with Salmonella [36, 55]. In addition to the ROS pathway, we also observed enrichment in the peroxisome pathway at early time point of HO53 treatment. Recently, Kim et al. [39] demonstrated an essential role of peroxisome proliferator-activated receptor α in innate host defense against Mtb. They showed that stimulation of peroxisome proliferator-activated receptor α led to activation of TFEB and increased lipid catabolism, depriving bacteria from the lipid source necessary for creation of an intracellular niche for Mtb replication [39]. Therefore, the interesting effect of HO53 on bronchial epithelium connected to enrichment in ROS and peroxisome pathways warrants further research on exploring APD compounds as potential modulators of PPARs. This concept gains further support as HO53 affected the fatty-acid metabolism pathway that in turn can regulate PPARs controlling lipid metabolism and inflammation [33]. The role of nuclear receptors such as PPARs or the xenobiotic nuclear receptor pregnane X receptor linked to drug metabolism has been shown to modulate the antimicrobial response against Mtb [35, 38]. In our model, HO53 affected the xenobiotic metabolism pathway in mature BCi cells, which might be linked directly to the metabolic turnover of HO53 but also to the observed promotion of autophagy.
Importantly, we demonstrated that HO53 activated the AMPK pathway together with enrichment in positively correlated glycolysis and negatively correlated OXPHOS pathways. Activation of AMPK can stimulate glycolysis in response to the energetic deficit in the cell and directly inhibits the mTOR pathway [36]. Interestingly, HO53 treatment led to activation of AMPK, enhancement of the glycolytic pathway, and inhibition of mTORC1 after 8 h that can explain the signaling cascade leading to autophagy induction at later time point. Thus, the enzyme catalyzing the first reaction of glycolysis, hexokinase-II, can directly bind and inhibit mTORC1, leading to autophagy induction [34]. This might explain inhibition of the mTORC1 pathway without enrichment in the glycolytic pathway observed after 24 h. Moreover, it has been demonstrated that activation of AMPK and enhanced expression of peroxisome proliferator-activated receptor-gamma coactivator 1α contribute to the autophagic clearance of Mtb and induced expression of genes involved in the OXPHOS [35]. In our model, we observed inhibition of OXPHOS during treatment with HO53 that might indicate additional regulatory mechanisms other than the AMPK-peroxisome proliferator-activated receptor-gamma coactivator 1α pathway.
In connection to the possible role of galectins in HO53-induced autophagy, we anticipated that HO53 increases production of galectins, which enhances detection of unidentified trigger/triggers in our model and activates AMPK, similar to the mechanism described previously, where galectin 9 recognized endolysosomal damage and activated AMPK [42]. We observed only slight increase in the galectin 9 protein level, ruling out our hypothesis that HO53 treatment activates the AMPK signaling cascade through increased galectin 9 protein expression and indicating additional mechanisms of AMPK activation. Further, the indication of mTOR inhibition by HO53 in airway epithelial cells was followed by investigation of TFEB nuclear translocation. TFEB is a key regulator of host innate immunity response to infections [56]. It has been shown to be responsible for the defense response against Staphylococcus aureus infections in mouse and in Caenorhabditis elegans by its ortholog HLH-30 [56]. In our model, translocation of TFEB to the nucleus is a rather late event linked to potential mechanism of autophagy induction by HO53 in mature bronchial epithelial cells. Despite of nonsignificant decrease in phosphorylation of S6K1, transcriptome analysis and the cellular localization of TFEB suggest that mTOR inhibition might contribute to the alterations in expression of autophagy-related ATG and MAP1LC3 genes. These genes encode proteins involved in formation of autophagosomes, and the enhanced expression of these genes corresponds to increased formation of autophagosomes analyzed by the LC3B marker and observed by TEM. Since TFEB translocated to the nucleus between 8 and 24 h and we observed changes in the expression of autophagy-related genes at earlier time points (4 h and 8 h), we conclude that additional regulatory mechanisms are most likely involved in induction of autophagy by HO53.
Treatment with HO53 led to a broad response and caused pronounced transcriptional changes in mature BCi cells. Moreover, the HO53 compound is related to the parental compound entinostat [57, 58], the histone deacetylase inhibitor [59], which led us to the hypothesis that mechanisms related to epigenetic regulation of the chromatin state could be involved in the HO53-induced autophagy. It has been shown that several histone-modifying epigenetic enzymes, e.g., G9a, EZH2, USP44, or CARM1 (coactivator-associated arginine methyltransferase 1) are important in the modulation of autophagy [50]. Interestingly, in our model, HO53 affected the most expression of genes encoding G9a − H3K9 methyltransferase, EZH2 − H3K27 methyltransferase, and USP44 − H2BK120 deubiquitinase. We anticipated that changes in the expression of histone-modifying enzymes might contribute to autophagy induction by decreased H2BK120Ub status as a result of increased USP44 expression and reduced H3K27me3 status caused by downregulation of EZH2 expression. We observed decreased ubiquitination of H2BK120 at 8 h followed by increased expression of USP44 at 24 h of HO53 treatment. Despite of changes in expression of EZH2, we did not observe any changes in H3K27me3 status. In mature BCi cells, observed epigenetic changes of histone modification and expression of histone-modifying enzymes were rather late events. However, regulation of the chromatin state is recognized as a dynamic process, and therefore, we would expect these changes at early time points.
When we consider the overall picture of molecular signaling caused by HO53, we can conclude that two regulatory mechanisms are intersected mainly at 8 h and 24 h time points. Hence, we suggest the mechanism for induction of autophagy by HO53 in mature bronchial epithelial cells as follows; HO53 treatment causes activation of AMPK (at 8 h) concomitant with decreased ubiquitination of H2BK120, followed by nuclear translocation of TFEB and increased expression of USP44 at 24 h. The mechanism of HO53-induced autophagy in mature human airway epithelial cells presents a complex picture, involving nuclear events that interplay with defined autophagy-related pathways, such as mTOR or AMPK. Our results and suggestion are not a complete version of HO53 mechanism, and additional effectors are most likely involved in the signaling cascade, leading to autophagy induction. Further investigation and evaluation of these additional effectors remain a future challenge as utilization of differentiated lung cells in the ALI culture has limitations in respect to genetic manipulations.
In summary, our results present a novel effect of the drug candidate HO53, which represents APD compounds and can be utilized as an inducer of autophagy in mature bronchial epithelial cells. Our studies focus on dissecting molecular pathways responsible for stimulation of autophagy by HO53 in mature bronchial epithelial cells. Although, the complete mechanism has not been resolved yet, our results warrant further studies including infections and in vivo models. Together with previously shown properties of HO53, enhancing epithelial barrier integrity, and production of AMPs, the compound has therapeutic potential for prevention and/or treatment of infections, limiting the usage of antibiotics and reducing the selection of antibiotic-resistant strains.
Statement of Ethics
Ethic approval of this study was not required as the worked is based on cell lines.
Conflict of Interest Statement
G.H.G. and B.A. are founders and stockholders in Akthelia Pharmaceuticals that holds a patent on APD compounds. Akthelia Pharmaceuticals was the patent applicant, and inventors are Roger Strömberg, Hakan Ottoson, Birgitta Agerberth, Gudmundur Gudmundsson, Erica Miraglia, and Frank Nylen (patent no. US 9,957,226 B2; status of application: patent is granted in the USA and in the EU). The APD compound HO53 used in this study is included in the patent as an inducer of innate immunity against infections. I.T.M., S.S., A.R.V., E.-L.E., and M.H.O declare no potential conflict of interest.
Funding Sources
This work was supported by the Icelandic Center for Research (RANNÍS) and University of Iceland research fund.
Author Contributions
I.T.M. designed and performed experiments, analyzed data, wrote, and edited the manuscript. S.S. performed initial analysis of the RNAseq data, A.R.V. performed experiments with VA10 cells, and E-L.E. analyzed TEM images. B.A. provided technical advises and together with S.S., A.R.V., and E-L.E. edited the manuscript. G.H.G. and M.H.O. contributed to the design of the experiments, interpretation of the data, and edition of the manuscript. All the authors reviewed and approved the final manuscript.
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on a reasonable request.
Supplementary Material
Supplementary data
Acknowledgments
We acknowledge deCODE genetics for the RNAseq and Arnar Pálsson for help with the experimental design and Sævar Ingþórsson, Jón Pétur Jóelsson, and Sigurdur Runar Gudmundsson for help with confocal microscopy and ImageJ analyses. We thank Paulina Cherek and Jóhann Arnfinnsson for sample preparation and image acquisition by TEM. We acknowledge Prof. Ronald G. Crystal and collaborators for generously providing us with the BCi-NS1.1 cell line. We thank Jennifer Kricker and Bryndis Valdimarsdottir for help with culture of BCi cells and Roger Strömberg and Hakan Ottosson for providing the HO53 compound for initial experiments.
References
- 1.Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018 Jun;19((6)):349–64. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011 Nov;147((4)):728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 3.Randow F, Münz C. Autophagy in the regulation of pathogen replication and adaptive immunity. Trends Immunol. 2012 Oct;33((10)):475–87. doi: 10.1016/j.it.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinsella RL, Nehls EM, Stallings CL. Roles for autophagy proteins in immunity and host defense. Vet Pathol. 2018 May;55((3)):366–73. doi: 10.1177/0300985818754967. [DOI] [PubMed] [Google Scholar]
- 5.Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019 Jan;176((1–2)):11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leiva-Juárez MM, Kolls JK, Evans SE. Lung epithelial cells: therapeutically inducible effectors of antimicrobial defense. Mucosal Immunol. 2018 Jan;11((1)):21–34. doi: 10.1038/mi.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bansil R, Turner BS. The biology of mucus: composition, synthesis and organization. Adv Drug Deliv Rev. 2018 Jan;124:3–15. doi: 10.1016/j.addr.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 8.Knowles MR, Boucher RC. Mucus clearance as a primary innate defense mechanism for mammalian airways. J Clin Invest. 2002 Mar;109((5)):571–7. doi: 10.1172/JCI15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004 Nov;306((5698)):1037–40. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 10.Gomes LC, Dikic I. Autophagy in antimicrobial immunity. Mol Cell. 2014 Apr;54((2)):224–33. doi: 10.1016/j.molcel.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Günther J, Seyfert HM. The first line of defence: insights into mechanisms and relevance of phagocytosis in epithelial cells. Semin Immunopathol. 2018 Nov;40((6)):555–65. doi: 10.1007/s00281-018-0701-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Junkins RD, Shen A, Rosen K, McCormick C, Lin TJ. Autophagy enhances bacterial clearance during P. aeruginosa lung infection. PLoS One. 2013 Aug;8((8)):e72263. doi: 10.1371/journal.pone.0072263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capasso D, Pepe MV, Rossello J, Lepanto P, Arias P, Salzman V, et al. Elimination of pseudomonas aeruginosa through efferocytosis upon binding to apoptotic cells. PLoS Pathog. 2016 Dec;12((12)):e1006068. doi: 10.1371/journal.ppat.1006068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vergne I, Chua J, Deretic V. Tuberculosis toxin blocking phagosome maturation inhibits a novel Ca2+/calmodulin-PI3K hVPS34 cascade. J Exp Med. 2003 Aug;198((4)):653–9. doi: 10.1084/jem.20030527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim YS, Silwal P, Kim SY, Yoshimori T, Jo EK. Autophagy-activating strategies to promote innate defense against mycobacteria. Exp Mol Med. 2019 Dec;51((12)):151. doi: 10.1038/s12276-019-0290-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rekha RS, Rao Muvva SSVJ, Wan M, Raqib R, Bergman P, Brighenti S, et al. Phenylbutyrate induces LL-37-dependent autophagy and intracellular killing of mycobacterium tuberculosis in human macrophages. Autophagy. 2015 Sep;11((9)):1688–99. doi: 10.1080/15548627.2015.1075110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuk J-M, Shin D-M, Lee H-M, Yang C-S, Jin HS, Kim K-K, et al. Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe. 2009 Sep;6((3)):231–43. doi: 10.1016/j.chom.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Mily A, Rekha RS, Kamal SMM, Arifuzzaman ASM, Rahim Z, Khan L, et al. Significant effects of oral phenylbutyrate and vitamin d3 adjunctive therapy in pulmonary tuberculosis: a randomized controlled trial. PLoS One. 2015 Sep;10((9)):e0138340. doi: 10.1371/journal.pone.0138340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bekele A, Gebreselassie N, Ashenafi S, Kassa E, Aseffa G, Amogne W, et al. Daily adjunctive therapy with vitamin D 3 and phenylbutyrate supports clinical recovery from pulmonary tuberculosis: a randomized controlled trial in Ethiopia. J Intern Med. 2018 Sep;284((3)):292–306. doi: 10.1111/joim.12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity. 2019 Feb;50((2)):432–45.e7. doi: 10.1016/j.immuni.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myszor IT, Parveen Z, Ottosson H, Bergman P, Agerberth B, Strömberg R, et al. Novel aroylated phenylenediamine compounds enhance antimicrobial defense and maintain airway epithelial barrier integrity. Sci Rep. 2019 Dec;9((1)):7114. doi: 10.1038/s41598-019-43350-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halldorsson S, Asgrimsson V, Axelsson I, Gudmundsson GH, Steinarsdottir M, Baldursson O, et al. Differentiation potential of a basal epithelial cell line established from human bronchial explant. Vitr Cell Dev Biol Anim. 2007 Oct;43((8–9)):283–9. doi: 10.1007/s11626-007-9050-4. [DOI] [PubMed] [Google Scholar]
- 23.Walters MS, Gomi K, Ashbridge B, Moore MS, Arbelaez V, Heldrich J, et al. Generation of a human airway epithelium derived basal cell line with multipotent differentiation capacity. Respir Res. 2013 Dec;14:135. doi: 10.1186/1465-9921-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C[T]) Method. Methods. 2001 Dec;25((4)):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Smith D, Helgason H, Sulem P, Bjornsdottir US, Lim AC, Sveinbjornsson G, et al. A rare IL33 loss-of-function mutation reduces blood eosinophil counts and protects from asthma. PLoS Genet. 2017 Mar;13((3)):e1006659. doi: 10.1371/journal.pgen.1006659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bjornsson E, Helgason H, Halldorsson G, Helgadottir A, Gylfason A, Kehr B, et al. A rare splice donor mutation in the haptoglobin gene associates with blood lipid levels and coronary artery disease. Hum Mol Genet. 2017 Jun;26((12)):2364–76. doi: 10.1093/hmg/ddx123. [DOI] [PubMed] [Google Scholar]
- 27.Bray NL, Pimentel H, Melsted P, Pachter L. Erratum: near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016 May;34((5)):525–7. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 28.Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, et al. Ensembl 2016. Nucleic Acids Res. 2016 Jan;44((D1)):D710–6. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. 2017 Jun;14((7)):687–90. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005 Oct;102((43)):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Möller K, Sigurbjornsdottir S, Arnthorsson AO, Pogenberg V, Dilshat R, Fock V, et al. MITF has a central role in regulating starvation-induced autophagy in melanoma. Sci Rep. 2019 Dec;9((1)):1–12. doi: 10.1038/s41598-018-37522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshii SR, Mizushima N. Monitoring and measuring autophagy. Int J Mol Sci. 2017 Sep;18((9)):1865. doi: 10.3390/ijms18091865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varga T, Czimmerer Z, Nagy L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim Biophys Acta. 2011 Aug;1812((8)):1007–22. doi: 10.1016/j.bbadis.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts DJ, Tan-Sah VP, Ding EY, Smith JM, Miyamoto S. Hexokinase-II positively regulates glucose starvation-induced autophagy through torc1 inhibition. Mol Cell. 2014 Feb;53:521–33. doi: 10.1016/j.molcel.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang CS, Kim JJ, Lee HM, Jin HS, Lee SH, Park JH, et al. The AMPK-PPARGC1A pathway is required for antimicrobial host defense through activation of autophagy. Autophagy. 2014 May;10((5)):785–802. doi: 10.4161/auto.28072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015 Mar;22((3)):377–88. doi: 10.1038/cdd.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.You L, Wang Z, Li H, Shou J, Jing Z, Xie J, et al. The role of STAT3 in autophagy. Autophagy. 2015 Jan;11((5)):729–39. doi: 10.1080/15548627.2015.1017192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhagyaraj E, Nanduri R, Saini A, Dkhar HK, Ahuja N, Chandra V, et al. Human xenobiotic nuclear receptor PXR augments mycobacterium tuberculosis survival. J Immunol. 2016 Jul;197((1)):244–55. doi: 10.4049/jimmunol.1600203. [DOI] [PubMed] [Google Scholar]
- 39.Kim YS, Lee H-M, Kim JK, Yang C-S, Kim TS, Jung M, et al. PPAR-α activation mediates innate host defense through induction of TFEB and lipid catabolism. J Immunol. 2017 Apr;198((8)):3283–95. doi: 10.4049/jimmunol.1601920. [DOI] [PubMed] [Google Scholar]
- 40.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017 Mar;168((6)):960–76. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thurston TL, Wandel MP, Von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012 Feb;482((7385)):414–8. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia J, Abudu YP, Claude-Taupin A, Gu Y, Kumar S, Choi SW, et al. Galectins control mTOR in response to endomembrane damage. Mol Cell. 2018 Apr;70((1)):120–35.e8. doi: 10.1016/j.molcel.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jia J, Abudu YP, Claude-Taupin A, Gu Y, Kumar S, Choi SW, et al. Galectins control MTOR and AMPK in response to lysosomal damage to induce autophagy. Autophagy. 2019 Jan;15((1)):169–71. doi: 10.1080/15548627.2018.1505155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012 Jun;5((228)):ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irazoqui JE. Key roles of MiT transcription factors in innate immunity and inflammation. Trends Immunol. 2020 Feb;41((2)):157–71. doi: 10.1016/j.it.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klionsky DJ, Cregg JM, Dunn WA, Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003 Oct;5:539–45. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 47.Yates B, Braschi B, Gray KA, Seal RL, Tweedie S, Bruford EA. Genenames.org: the HGNC and VGNC resources in 2017. Nucleic Acids Res. 2017 Jan;45((D1)):D619–25. doi: 10.1093/nar/gkw1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.HUGO Gene Nomenclature Committee (HGNC), European Molecular Biology Laboratory. Hinxton, Cambridge CB10 1SD, United Kingdom: European Bioinformatics Institute (EMBL-EBI), Wellcome Genome Campus; https://www.genenames.org/ [Google Scholar]
- 49.Gammoh N, Marks PA, Jiang X. Curbing autophagy and histone deacetylases to kill cancer cells. Autophagy. 2012 Oct;8((10)):1521–2. doi: 10.4161/auto.21151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baek SH, Kim KI. Epigenetic control of autophagy: nuclear events gain more attention. Mol Cell. 2017 Mar;65((5)):781–5. doi: 10.1016/j.molcel.2016.12.027. [DOI] [PubMed] [Google Scholar]
- 51.Chen S, Jing Y, Kang X, Yang L, Wang DL, Zhang W, et al. Histone H2B monoubiquitination is a critical epigenetic switch for the regulation of autophagy. Nucleic Acids Res. 2017 Feb;45((3)):1144–58. doi: 10.1093/nar/gkw1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Artal-Martinez de Narvajas A, Gomez TS, Zhang J-S, Mann AO, Taoda Y, Gorman JA, et al. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol. 2013 Oct;33((20)):3983–93. doi: 10.1128/MCB.00813-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Losier TT, Akuma M, McKee-Muir OC, LeBlond ND, Suk Y, Alsaadi RM, et al. AMPK promotes xenophagy through priming of autophagic kinases upon detection of bacterial outer membrane vesicles. Cell Rep. 2019 Feb;26((8)):2150–65.e5. doi: 10.1016/j.celrep.2019.01.062. [DOI] [PubMed] [Google Scholar]
- 54.Shen S, Niso-Santano M, Adjemian S, Takehara T, Malik SA, Minoux H, et al. Cytoplasmic STAT3 represses autophagy by inhibiting PKR activity. Mol Cell. 2012 Dec;48((5)):667–80. doi: 10.1016/j.molcel.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 55.Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MAO, et al. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci USA. 2009 Apr;106((15)):6226–31. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves AMF, Wollenberg AC, et al. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity. 2014 Jun;40((6)):896–909. doi: 10.1016/j.immuni.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ottosson H, Nylén F, Sarker P, Miraglia E, Bergman P, Gudmundsson GH, et al. Potent inducers of endogenous antimicrobial peptides for host directed therapy of infections. Sci Rep. 2016 Nov;6:36692. doi: 10.1038/srep36692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miraglia E, Nylén F, Johansson K, Arnér E, Cebula M, Farmand S, et al. Entinostat up-regulates the CAMP gene encoding LL-37 via activation of STAT3 and HIF-1α transcription factors. Sci Rep. 2016 Sep;6:33274. doi: 10.1038/srep33274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knipstein J, Gore L. Entinostat for treatment of solid tumors and hematologic malignancies. Expert Opin Investig Drugs. 2011 Oct;20((10)):1455–67. doi: 10.1517/13543784.2011.613822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author on a reasonable request.