Summary
Several cross-protective antibodies that recognize a broad range of influenza A virus (IAV) strains are known to have functions in virus elimination such as Fcγ receptor (FcγR)-effector function and neutralizing activity against the head region. Although few studies have used primary cells as effector cells, the FcγR-effector function was evaluated after isolating each cell subset. Herein, we established an original assay system to evaluate purified FI6 IgG-mediated binding to hemagglutinin (HA)-expressing cells by flow cytometry using peripheral blood mononuclear cells from cynomolgus macaques. In addition, we evaluated the FcγR-effector function of IAV vaccine-induced anti-HA antibodies in cynomolgus macaques after administering the split vaccine. We found several cell types, mainly classical monocytes, bound to HA-expressing target cells in an FcγR-dependent manner, that were dominant in the binding of the cell population. Thus, this assay system could facilitate the development of a universal influenza vaccine.
Subject areas: Biological sciences, Immunology, Immunological methods
Graphical abstract

Highlights
-
•
A novel assay system for evaluation of FcγR-effector function in cynomolgus macaque PBMCs
-
•
Several different cell types bound to HA-expressing cells in the FcγR-dependent manner
-
•
IgGs elicited by flu vaccination induced FcγR-dependent classical monocytes binding
-
•
This assay system could facilitate the development of a universal influenza vaccine
Biological sciences; Immunology; Immunological methods
Introduction
Current vaccines against seasonal influenza A viruses (IAVs) protect against infection by inducing neutralizing antibodies against the immunodominant region, which is predominantly the head domain of hemagglutinin (HA). IAVs often exhibit antigenic drift mainly in the head region, and highly pathogenic avian IAVs are considered a threat because they can cause pandemics because of their sporadic transmission to humans and resultant high mortality rates (Subbarao, 2018). Therefore, the protection efficacy of vaccines is reduced against not only pandemic strains but also antigen-mismatched seasonal IAV strains (Nelson and Holmes, 2007). Accordingly, there is an urgent need to develop a universal influenza vaccine that can induce effective immunity against a broad range of influenza virus strains, including not only seasonal IAV strains but also pandemic strains.
Various approaches have been used to develop universal influenza vaccines. One of the major attempts is the identification of cross-protective antibodies against broad IAV strains and their application for vaccine development by identifying the antigen epitope’s regions recognized by these antibodies (Corti et al., 2017). In general, cross-protective antibodies demonstrate a broad spectrum of protection against infection by recognizing conserved epitopes that are poorly mutated. The candidate epitopes are the stem domain (Adachi et al., 2019; Corti et al., 2011; Tan et al., 2012), the receptor-binding site in the head domain (Shen et al., 2017; Whittle et al., 2011), and the lateral patch (Raymond et al., 2018) or vestigial esterase site (Bangaru et al., 2018). A broadly cross-reactive but non-neutralizing antibody targeting the trimer interface in the head domain has also been reported (Bangaru et al., 2019; Watanabe et al., 2019). These antibodies were mostly identified by the B-cell receptor sequences of B cells capable of recognizing a broad range of IAV strains in the blood after vaccination. In addition, one strategy could be to aim to provide this information to induce these cross-protective antibodies through vaccination, as has been reported in HIV (Jardine et al., 2015). Other attempts to design antigens have also been made. For example, it has been reported that immunization with HA bound to nanoparticles can induce cross-protective antibodies (Darricarrere et al., 2021; Kanekiyo et al., 2019).
To evaluate a developing vaccine, the hemagglutination inhibition (HAI) assay, the standard assay for estimating the efficacy of current IAV vaccines, is used for measuring the neutralizing activity against the influenza virus based on the binding capacity to epitopes around the receptor-binding site in the HA head region. However, it is not adequate to evaluate the overall function of antibodies that recognize a broad range of viral strains. Cross-protective antibodies are known to have Fcγ receptor (FcγR) effector function in defense against IAV infection in vivo in addition to neutralizing activity, and the development of a novel assay system is required (Adachi et al., 2019; Bournazos et al., 2020; DiLillo et al., 2014, 2016; Maamary et al., 2017).
There are three types of human Fc receptors, FcγRI (CD64), FcγRII (CD32), and FcγRIIIa (CD16), (Bruhns, 2012). In humans and monkeys, IgG1 and IgG3 readily bind to natural killer (NK) cells, neutrophils, monocytes, and macrophages, which express FcγRIIIa, whereas monocytes, macrophages, and dendritic cells express FcγRI and FcγRII (Jegaskanda et al., 2014; Mullarkey et al., 2016; Seidel et al., 2013) and activate cells via FcγR, leading to antibody-dependent cellular cytotoxicity (ADCC) and antibody-dependent cellular phagocytosis (ADCP) (Boudreau and Alter, 2019). Several assays have been reported to evaluate FcγR-effector functions, such as ADCC and ADCP activities for specific cells such as NK cells, macrophages, and monocytes (Ana-Sosa-Batiz et al., 2016; Simhadri et al., 2015; Vanderven et al., 2016). Most of these utilize cultured cell lines as effector cells, and the mode of FcγR expression differs from that of primary cells. Furthermore, the FcγR-effector function is involved in the activation of CD8+T cells in addition to NK cells, macrophages, monocytes, dendritic dells (Bournazos et al., 2020), and B cells (Maamary et al., 2017) rendering the evaluation of only specific cells as incomplete. Therefore, it is necessary to develop a system that can simultaneously evaluate the FcγR-effector function in primary cells. Moreover, for vaccine development, the selection of an animal model for vaccination is essential. The mouse is not a natural host for influenza, and influenza viruses that cause epidemics and pandemics in humans usually replicate inefficiently in the mouse unless an adapted virus is used. Furthermore, the subclasses of IgG and the expression pattern of FcR are known to differ from those in humans. Mice express a novel activating receptor, FcγRIV, which is functionally similar to human FcγRIIIa. However, mouse FcγRIV differs from human FcγRIIIa in distribution and IgG isotype binding (Bruhns, 2012). In mice, FcγRIV binds to IgG2a, IgG2b, and IgE, but not to IgG1 or IgG3. Non-human primate models such as rhesus and cynomolgus macaques were used to study the FcγR-effector function and showed that FcγR-mediated immune response-inducing antibodies are associated with reduced influenza virus shedding (Florek et al., 2014; Jegaskanda et al., 2013). Regarding FcγR, the distribution of FcγR expression is similar in humans and NHPs, making NHPs useful animal models for predicting FcγR function in humans. However, certain minor differences have been reported concerning the distribution of FcγR (Crowley and Ackerman, 2019). One of the differing aspects is the FcγRIIIb receptor, which contributes to the activation of granulocytes and is found on human granulocytes, while not present in macaque (Fossati et al., 2002).
In this study, we aimed to clarify the issues pertaining to the current vaccine by evaluating the antibodies induced by split vaccine administration in cynomolgus macaques and the associated FcγR-effector function in individual cells using peripheral blood mononuclear cells (PBMCs).
Results
Generation of HA-expressing target cells
To quantitatively monitor the relative number of target cells, we first developed a cell line stably expressing the luciferase gene. Because the natural target of IAV was lung epithelial cells, the human lung epithelial A549 cell line was chosen as the parental cell line for the target cells. A549 cells were transduced with the luciferase gene under the control of the mouse retroviral long terminal repeat promoter (A549-luc). Because original A549 cells undergo cell death due to the spontaneous killing activity of NK cells when co-cultured with them, we selected an A549 clone that is resistant to their antibody-independent cytotoxic activity. For this, the A549-luc cells were first co-cultured with an NK cell line, KHYG-1, for one week and then co-cultured with another NK cell line, NK92 mi, for an additional week (total two weeks; Figure S1). We then cloned A549-NKR-luc cells as parental cells to establish cell lines stably expressing IAV hemagglutinin of H1 A/Puerto Rico/8/1934 (PR8-HA) and H1 A/California/07/2009 (CA-HA) cells.
Next, to establish a sensitive assay system to monitor the FcγR-effector function in non-human primate (NHP) cells, we modified FI6, which is known as a broad-spectrum anti-HA neutralizing monoclonal antibody (Corti et al., 2011), into a cynomolgus-type Fc region. We then generated FI6-cynoIgG1 and FI6-cynoIgG2 and purified them. The binding of cynoIgGs to HA on the cell surface of HA-expressing A549-NKR-luc cells was analyzed by flow cytometry. Both FI6-cynoIgG1 and IgG2 but not control humanIgG1 bound to HA to the same extent and in a dose-dependent manner (Figure 1 A and B). Therefore, these cynoIgGs were utilized to evaluate the Fc-effector functions in the following study.
Figure 1.
Establishment of target cells
(A) Detection of CA-HA-expression and (B) IgG dose-dependent curve in A549-NKR-luc cells. HA expression was determined by flow cytometric analysis using each dose of FI6-cynoIgG1, IgG2, or control human IgG1 (n = 4, ∗p< 0.05 by Mann-Whitney U-test ). Data are represented as mean ± SD.
Establishment of antibody-dependent binding assay
To evaluate whether these cynoIgGs can enable the cross-linking of HA-expressing target cells with immune effector cells in an FcγR-dependent manner, we used CFSE-labeled PBMCs and performed quantitative analysis by flow cytometry. First, we used CA-HA-expressing cells as target cells and target-effector binding was evaluated by the CFSE-positive population in the forward scatter high large-cell subset (Figure 2A). The binding of FI6-cynoIgG1 to target cells significantly increased the frequency of PBMC binding in a dose-dependent manner compared to control human IgG1 treatment (Figures 2B and 2C).
Figure 2.
Establishment of an antibody-dependent binding assay
(A) Gating scheme for the binding assay. Detection of binding cells between CA-HA-expressing A549-NKR-luc cells and cynoPBMCs (B) and (C) IgG dose-dependency. Each concentration of FI6-cynoIgG1 was bound to HA-expressing cells mixed with CFSE-labeled cynoPBMCs and subjected to flow cytometric analysis (n = 8, ∗∗p< 0.01 by Wilcoxon matched-pairs signed rank test). The frequency of binding cells in (D) CA and (E) PR8-HA-expressing A549-NKR-luc cells and cynoPBMCs by each IgG subclass (n = 9, ∗∗p< 0.01 by Wilcoxon matched-pairs signed rank test). The frequency of binding cells in (F) CA and (G) PR8-HA-expressing A549-NKR-luc cells and cynoPBMCs by FI6-IgG1 and FI6-cynoIgG1 LALA. (n = 8, ∗∗p< 0.01 by Wilcoxon matched-pairs signed rank test).
Next, we examined whether the frequency of binding PBMCs bound was different for each IgG subclass. As a result, both FI6-cynoIgG1 and IgG2 significantly increased the frequency of binding cells compared to PBS treatment (Figure 2D). However, in a comparison of IgG subclasses, FI6-cynoIgG1 showed significantly more PBMCs than IgG2 (Figure 2D). The same trend was observed in PR8-HA-expressing cells (Figure 2E). To confirm that the binding of PBMCs to target cells depends on antibody-FcγRs interaction, we generated FI6-cynoIgG1 harboring amino acid mutations L234A and L235A (LALA), which lacks the ability to bind to FcγRs in human and macaques (Lund et al., 1991; Parsons et al., 2019). When FI6-cynoIgG1 LALA was used instead of parental IgG1, the frequency of binding cells dramatically decreased to the same degree as in PBS (Figures 2F and 2G), suggesting the requirement of Fc- FcγRs binding for the target-effector interaction observed in this assay. These results are consistent with previous reports from human IgG subclass Fc-effector function assays (deTaeye et al., 2020; Vafa et al., 2014), suggesting that this assay could be utilized as a sensitive antibody-dependent binding assay using NHP samples.
Imaging flow cytometric analysis of binding cells
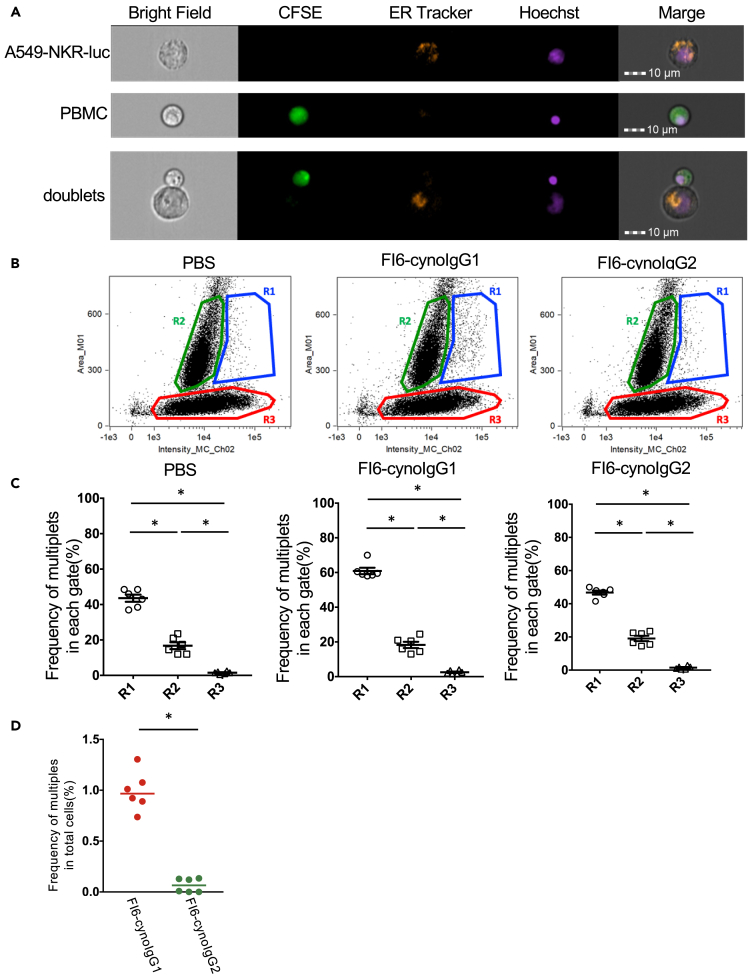
To confirm the FcγR-dependent target-effector binding, which was observed by conventional flow cytometry, we analyzed the binding using imaging flow cytometry, by which the multiplet status was visualized using fluorescent microscopy. To distinguish the target cells from effector PBMCs, A549-NKR-luc cells were first marked with orange fluorescent-conjugated ER-Tracker, then the ER-Tracker labeled target cells and CFSE-labeled PBMCs were incubated in the presence or absence of FI6-cynoIgGs. Using imaging flow cytometry, ER-Tracker labeled target cells and CFSE-labeled PBMCs were detected as either singlets or multiplets (Figure 3A). To visualize the multiplets on the scatter plot, plotting was performed with the intensity of CFSE (x-axis) and area (y-axis; an indicator of the cell size as FCS-A parameter in flow data) as in flow cytometry analysis (where target-effector doublets were defined by the FCS-A versus CFSE intensity). Consequently, the plot resembled that of flow cytometry (Figures 2A and 3B). For the quantification, the cells were divided into three regions (R1, R2, and R3). Region 1 (R1; large size and CFSE positive) indicated multiplets, Region 2 (R2; small size but CFSE positive) indicated CFSE-labeled PBMC singlet, and Region 3 (R3; large size but CFSE negative) indicated unbound target cells. To confirm whether R1 indeed contains the multiplets cells as the consequence of target-effector binding, we visually evaluated the mode of multiplets (Figure S2) in each region carefully. We then counted the multiplets for each region (Figures S2A and S2B) and found that the R1 had the most doublets in each region (Figure 3C). When the percentage of multiples was calculated for R1, the frequency of multiples was significantly higher in FI6-cynoIgG1 than in FI6-cynoIgG2 (Figure 3D). This result was similar to that of the binding assay. These results indicate that the doublets detected by binding assay are the aggregates of effector cells bound to the target cells via the antibody-FcγR interaction.
Figure 3.
Imaging flow cytometric analysis of binding cells
(A) Representative images by imaging cytometer analysis. CynoPBMCs were labeled with CFSE (green). A549-NKR-luc cells were labeled with ER tracker (orange). Both cells of nuclei were stained with Hoechst 33342 (violet).
(B) Representative plots by imaging cytometer analysis. Three distinct populations (R1, R2, and R3) were gated by expanding the intensity of CFSE and area.
(C) The frequency of multiplets in each region. After the binding assay, imaging analysis was performed (Figure S2) (n = 6, ∗p< 0.05 by Wilcoxon matched-pairs signed rank test). Data are represented as mean ± SEM.
(D) The frequency of multiplets in R1 of each IgG subclass (n = 6, ∗ p< 0.05 by Wilcoxon matched-pairs signed rank test). See also Figure S2.
FI6cynoIgG1 binds NK cells and classical monocytes to target cells via FcγRs
To investigate which immune cell subset in PBMCs is bound to the target cells, a staining panel for monkey PBMCs was designed to identify the expression of FcγRs and a subset of interest was created in the duplex gate (Figure S3A). As shown in Figure 4A, treatment with FI6-cynoIgG1 resulted in the significant increase in the number of CD16+CD32+CD64+, CD16+CD32−CD64−, CD16−CD32+CD64+, and CD16−CD32+CD64− cells compared to that in PBS treatment. In contrast, treatment with FI6-cynoIgG2 increased the number of CD16−CD32+CD64+ cells as well as FI6-cynoIgG1, and the number of CD16−CD32−CD64+ cells was significantly increased compared to that in PBS treatment, which suggests that there is a wide variation in target-effector binding. Each CD16/CD32/CD64 single positive cell subset suggested the involvement of all three types of FcγRs for target-effector binding. Next, we investigated the cell phenotype of PBMCs bound to CA-HA-expressing target cells. We found that the target binding immune cells were mainly NK cells and classical monocytes (Figures 4B and 4C). The same trend was observed for PR8-HA-expressing target cells (Figures S3B–S3D).
Figure 4.
FI6cynoIgG1 binds NK cells and classical monocytes to target cells via FcγRs
(A) The distribution of FcγR expression in binding cells. Flow cytometric analysis was conducted after the binding assay using CA-HA-expressing cells using subset markers and FcγRs antibodies (n = 7, ∗p< 0.05 by Wilcoxon matched-pairs signed rank test). The (B) number and (C) frequency of each subset in FcγR expression cells of FI6-cynoIgG1 binding. The number of each cell in FI6-cynoIgG1 treated was subtracted by that in PBS treated. Data are represented as mean ± SD. See also Figure S3.
Taken together, these results suggest that the FcγR-dependent effector function induced by purified cynoIgG1 is likely mediated by NK cells and monocytes.
HA split vaccine mediates Fc-effector function via classical monocytes
We developed and validated an assay system to simultaneously evaluate various types of FcγR-mediated target-effector binding by using target cells with purified FI6 monoclonal antibodies and CFSE-labeled PBMCs. To evaluate the potential application of this assay for vaccine development, we examined whether our assay can be useful for analyzing FcγR-mediated target-effector binding by using sera from cynomolgus monkeys vaccinated in vivo. To obtain vaccine-induced anti-HA serum, monkeys were administered twice with the HA split vaccine against IVA CA according to the standard prime-boost regimen, and their blood was collected over time (Figure S4A). Anti-HA antibody titers in the serum increased significantly after vaccination in all monkeys examined (Figure 5A). Next, we performed a binding assay using sera from the sixth week after vaccination by diluting them according to the measured antibody titer and matching the amount of antibody bound to the target cells. Serum IgG were found to bind to the CA-HA-expressing target cells (Figure S4B), although there was no correlation between the number of bound cells and antibody titer (Figure 5B). Finally, the target-effector binding was examined (Figure 2) and found that the number of CD16−CD32+CD64+, CD16+CD32−CD64+, CD16−CD32+CD64−, and CD16−CD32−CD64+ cells significantly increased compared to that in PBS treatment, as observed in the case of FI6 (Figure 5C).
Figure 5.
HA split vaccine mediates Fc effector function via classical monocytes
(A) Total IgG antibody titers against CA-HA after split vaccine immunization in cynomolgus monkeys (n = 6).
(B) The correlation between anti-CA-HA titer at six weeks and the number of binding cells (by Spearman correlation test).
(C) The distribution of FcγR expression in binding cells. After the binding assay using CA-HA-expressing cells, flow cytometric analysis was conducted using anti-subset markers and FcγRs antibodies (n = 6, ∗p< 0.05 by Wilcoxon matched-pairs signed rank test).
(D) The number and (E) frequency of each subset in FcγR expression cells of serum at 6 weeks. The number of each cell in serum treated was subtracted by that in PBS treated. Data are represented as mean ± SD.
(F) The number and (G) frequency of each subset in FcγR expression cells of purified IgG in serum at 6 weeks. The number of each cell in purified IgG serum treated was subtracted by that in PBS treated. Data are represented as mean ± SD. See also Figure S4.
Unlike FI6, the number of CD16+CD32−CD64− cells was not increased by serum treatment. Coincidently, the subset of cells expressing FcγR was mostly classical monocytes (Figures 5D and 5E). These results suggest that, at least in cynomolgus monkeys, the FcγR-effector function induced by the serum of the HA split vaccine is mainly mediated by classical monocytes.
To examine the involvement of FcRs other than FcγR, IgG in serum was purified and the binding assay was performed. The purified IgG in serum treatment showed the same trend as that of serum (Figures 5F and 5G), which suggested that the binding PBMCs and target cells occurred mainly via anti-HA specific IgG. Furthermore, to examine the activation status of the binding classical monocytes, we assessed the induction of activation markers in the binding classical monocytes after binding to HA-CA target cells compared to that in unbound cells after serum treatment. The expression of CD80 mRNA was significantly upregulated, and CD83 mRNA was trended to be upregulated in the binding classical monocytes. It suggests that the classical monocytes were activated after binding to the target cells via anti-HA specific IgG (Figure S5A).
Discussion
In this study, we aimed to establish a PBMC-based assay system to evaluate FcγR-mediated target-effector binding, which potentially reflects the effector function of FcγR. The effector function of FcγR is critical for the development of a universal influenza vaccine. Most of the studies on ADCC and/or ADCP have used cell lines as the effector cells, and the presence or absence of cell–cell interaction has been ignored in cell-line based assays. However, in reality, there are multiple cells expressing FcγRs, and the effector cells exist as heterogeneous cell populations in the body. Thus, it is difficult to fully understand FcγR-mediated immune responses using cell lines. Therefore, we thought it was worth establishing an assay based on PBMCs, including multiple different immune cells, instead of a cell line.
However, it is advantageous to use the NHP model as an animal model to estimate the efficacy of vaccines, and the in vivo response after vaccine or antibody administration can be confirmed. Another important point in the use of NHP is that the expression distribution of FcγR is similar to that in humans. However, there has been no comprehensive assay system for the FcγR-effector function using the NHP model. Because we tested the system’s applicability to non-clinical studies using the serum after administration of the H1N1 split influenza vaccine in cynomolgus macaques, the present study could be the first report to overcome these issues.
IgG1 is known to have a higher affinity than IgG2 for binding to each FcγR (Bruhns, 2012), and FI6-cynoIgG1 predominantly induces binding of CD16+CD32−CD64−, CD16−CD32+CD64+, and CD16+CD32+CD64+ cells to HA-expressing target cells. The binding of CD16+CD32−CD64− cells to target cells is likely to occur via CD16. For CD16−CD32+CD64+ and CD16+CD32+CD64+ cells, binding to target cells is expected to occur mainly through CD64 because it has a higher binding capacity to IgG1 than CD32 and CD16, Therefore, all expressed FcγRs are functional for the binding. Furthermore, FI6-cynoIgG1-mediated binding utilizes NK cells and classical monocytes. However, NK cells may be included in CD16+CD32−CD64− and classical monocytes in CD16−CD32+CD64+ cell populations. Thus, it is clear that a purified antibody does not induce a single cell type; instead, multiple cell types bind to the target cells via the antibody and induce the FcγR-effector function. Although Expi293 was used to produce the recombinant FI6-cynoIgG used in this study, it is known that glycosylation differs depending on the type of cell line from which recombinant IgGs are produced. Glycosylation variation results in changes in the binding affinity and functionality of IgGs with FcRs (Boyoglu-Barnum et al., 2020; He et al., 2016; Jennewein and Alter, 2017). Therefore, it is unclear whether the glycosylation of the antibody in vivo can be directly reflected in this assay. Moreover, it has also been reported that ADCC induction is inhibited by high-titer HAI antibodies or the addition of an antibody with HAI activity to the serum of vaccines (Cox et al., 2016). Therefore, it is necessary to comprehensively evaluate antibodies in the serum induced by vaccine administration.
To comprehensively evaluate the antibodies that bind to HA in serum, we evaluated the FcγR-effector function of the current seasonal influenza virus vaccine using sera obtained after two doses of A/CA/07/2009 split vaccine in cynomolgus macaques. Because there was no correlation between the antibody titer and the number of PBMCs bound to the target cells, it is difficult to predict the FcγR-effector function based on the quantity of HA-specific IgG. NK cells were not well utilized as FcγR-mediated bound partners in sera of vaccinated cynomolgus macaques, unlike when purified FI6-cynoIgG1 was treated. Previous reports have shown that seasonal influenza vaccination does not sufficiently induce broadly reactive ADCC-induced antibodies, which is similar to the results of this study and suggests the usefulness of this evaluation system (Jacobsen et al., 2017; Jegaskanda et al., 2016; Vanderven et al., 2017). It has been reported that anti-influenza virus IgG monoclonal antibodies selectively bind to activated FcγRIIa to promote dendritic cell maturation and induction of CD8+T cell responses (Bournazos et al., 2020), IgG produced by the inoculation may be involved in the FcγR-effector function via conventional dendritic cells, enhancing the efficacy of prevention and treatment of IAV infection.
It has been reported that trivalent influenza vaccination increases ADCP activity against the HA protein of the vaccine strain in adults (Ana-Sosa-Batiz et al., 2017). THP-1 cells, a monocyte cell line, were used in this study. We consistently demonstrated the involvement of monocytes in FcγR-mediated target-effector binding. Furthermore, detailed classification of monocytes revealed that among the three monocyte subsets, classical monocytes were mainly involved in binding. Because classical monocytes have been reported to have a higher phagocytic capacity than other monocyte subsets (Mukherjee et al., 2015), split vaccine administration is expected to bind classical monocytes to target cells via induced antibodies and exhibit ADCP activity. However, the level was the same as that of the treatment with FI6-cynoIgG1 purified antibody, which may be the limit of the ability of IgG to bind classical monocytes. Based on the above, we believe that ADCC activity can be enhanced by adding a Th1-type adjuvant that can induce NK cells to the current influenza split vaccine by adding purified antibody with high FcγR-effector activity or by changing the design of the antigen. This will lead to the development of a universal influenza vaccine. In addition, cross-reactivity with a broader range of HA strains could also be evaluated by changing the HA expressed by the target cells.
In conclusion, we developed an assay system to evaluate the FcγR-effector function using cynomolgus macaque PBMCs as effector cells and verified the effect using serum and PBMCs after split vaccine administration. The results showed that the antibodies induced by the split vaccine mediate FcR-dependent target-effector binding, and the main subset of the bound cell is classical monocytes. This assay system will be helpful for the development of a universal influenza vaccine because it will allow us to examine in detail the FcγR-effector function of individual cells induced by antibodies from passive immunization or vaccine administration in NHPs.
Limitations of the study
Because we analysis the binding activity in PBMCs in this study, we were unable to evaluate the FcγR effector function of blood cells such as neutrophils and other granulocytes and FcγR bearing cell in tissue like alveolar macrophages (He et al., 2017). We did not examine whether effector cells bound to the target cells are involved in the cytotoxicity, or phagocytosis of the target cells, and it will be essential to examine the function of effector cells in the future. In addition, as there is an inhibitory CD32B subtype of CD32, it is necessary to examine whether all bound cells have the FcγR-effector function.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-human CD64-PE (Clone 10.1) | BD Biosciences | Cat.# 558592; RRID: AB_647202 |
| Mouse anti-human CD123-Cy7PE (Clone 7G3) | BD Biosciences | Cat.# 560826; RRID: AB_10563898 |
| Mouse anti-human CD56-Alexa Fluor 700 (Clone B159) | BD Biosciences | Cat.# 561902; RRID: AB_10894388 |
| Mouse anti-human CD32-BV786 (Clone FL18.26) | BD Biosciences | Cat.# 564840; RRID: AB_2738978 |
| Mouse anti-human CD3-BUV615 (Clone SP-34) | BD Biosciences | Cat.# 751249; RRID: AB_2875266 |
| Mouse anti-human CD14-BUV805 (Clone M5E2) | BD Biosciences | Cat.# 565779; RRID: AB_2716868 |
| Mouse anti-human CD11c-PE-Cy5 (Clone 3.9) | BioLegend | Cat.# 301610; RRID: AB_493578 |
| Mouse anti-human CD16-APC-Cy7 (Clone 3G8) | BioLegend | Cat.# 302018; RRID: AB_314218 |
| Mouse anti-human CD20-BV711 (Clone 2H7) | BioLegend | Cat.# 302342; RRID: AB_2562602 |
| Mouse anti-human CD159a (NKG2A)-APC (Clone Z199) | Beckman Coulter | Cat.# A60797; RRID: AB_10643105 |
| Mouse anti-human HLA-DR–PE-Texas Red (Clone TU36) | Life Technologies | Cat.# MHLDR17; RRID: AB_10392851 |
| Anti-FI6-cynoIgG1 | This paper | N/A |
| Anti-FI6-cynoIgG2 | This paper | N/A |
| control human IgG1 (Clone QA16A12) | BioLegend | Cat.# 403502; RRID: AB_2847831 |
| biotin-conjugated anti-human total IgG antibody(Clone G18-145) | BD Biosciences | Cat.# 555785; RRID: AB_396120 |
| Goat anti-human IgG (H + L) Cross-Adsorbed Secondary Antibody-Alexa Fluor 647 | Thermo Fisher Scientific | Cat.# A-21445; RRID: AB_2535862 |
| Biological samples | ||
| Peripheral blood mononuclear cells from cynomolgus macaques | Tsukuba Primate Research Center, National Institutes of Biomedical Innovation, Health and Nutrition | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| CFSE | Thermo Fisher Scientific | Cat.# C34554 |
| ER tracker Red | Thermo Fisher Scientific | Cat.# E34250 |
| Hoechst 33342 | Thermo Fisher Scientific | Cat.# H3570 |
| HRP-conjugated streptavidin | Thermo Fisher Scientific | Cat.# N100 |
| Critical commercial assays | ||
| THUNDERBIRD SYBR qPCR Mix | TOYOBO | Cat.# QPS-201 |
| RNeasy RNeasy MicroKit | QIAGEN | Cat.# 74004 |
| SuperScript III First-Strand Synthesis System | Thermo Fisher Scientific | Cat.# 18080051 |
| Experimental models: Cell lines | ||
| Platinum-A cells | Cell Biolabs, Inc. | Cat.# RV-102 |
| A549 cells | JCRB | Cat.# JCRB0076 |
| NK92mi cells | ATCC | Cat.# CRL-2408 |
| KHYG-1 cells | JCRB | Cat.# JCRB0156 |
| Expi293F cells | Thermo Fisher Scientific | Cat.# A14527 |
| Experimental models: Organisms/strains | ||
| Cynomolgus macaques | Tsukuba Primate Research Center, National Institutes of Biomedical Innovation, Health and Nutrition | N/A |
| Oligonucleotides | ||
| Forward primer for cynoGAPDH 5′-AGAAGTATGACAACAGCCTCA-3′ |
This paper | N/A |
| Reverse primer for cynoGAPDH 5′-ACTGTGGTCATGAGTCCTCC-3′ |
This paper | N/A |
| Forward primer for cynoCD80 5′-AAACTCGCATCTACTGGCAAA-3′ |
This paper | N/A |
| Reverse primer for cynoCD80 5′-GGTTCTTGTACTCGGGCCATA-3′ |
This paper | N/A |
| Forward primer for cynoCD83 5′-AAAGCTGGCATGGAACGAG-3′ |
This paper | N/A |
| Reverse primer for cynoCD83 5′-TGTCTTGTGAAGAGTCACTGGC-3′ |
This paper | N/A |
| Recombinant DNA | ||
| pMx-IRES-Puro vector | Cell Biolabs, Inc. | Cat.# RTV-014 |
| pCMV/R H1 A/Puerto Rico/8/1934 (PR8) vector | This paper | GenBank: NP_040980 |
| pCMV/R H1 A/California/07/2009 (CA) vector | This paper | GenBank: FJ969540 |
| pcDNA3.1-FI6 cynoIgG1 heavy chain vector | This paper | GenBank: JN234439 |
| pcDNA3.1-FI6 cynoIgG2 heavy chain vector | This paper | GenBank: CS101554 |
| pcDNA3.1-FI6 light chain vectors | This paper | GenBank: JN234448 |
| Software and algorithms | ||
| FlowJo 10.7.1 | BD Biosciences | https://www.flowjo.com/ |
| IDEAS software 6.2 | Luminex | https://www.luminexcorp.com |
| Prism 7.0a | GraphPad | https://www.graphpad.com |
| SPICE 6.0 | Roederer et al., 2011 | https://niaid.github.io/spice/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Takuya Yamamoto (yamamotot2@nibiohn.go.jp).
Materials availability
Newly generated plasmids, antibodies, and genetically modified cell lines in this study are available from the lead contact upon request.
Experimental model and subject details
Animal experiments and sampling of PBMCs
In total, five male and fifteen female cynomolgus macaques aged 4–10 years were maintained and used in this study. We did not find a significant influence on both sexes in this study. All animals were maintained in accordance with the Guide for the Care and Use of Laboratory Animals, National Institutes of Health Report no. 85-23 (Department of Health and Human Services, Bethesda, MD, 1985). All animals were housed in the Tsukuba Primate Research Center of the National Institute of Biomedical Innovation after full protocol approval from the Institutional Animal Care (DS30-41R1). Two male and four female cynomolgus macaques aged 5–8 years were intramuscularly vaccinated twice with 15μgA/CA/07/2009 H1N1 split vaccine at 0 and 3 weeks. Blood samples were collected weekly starting from 0 week. In addition, sample collection, PBMCs, and plasma isolation from blood samples containing EDTA by Ficoll-Paque PLUS (Cytiva) was performed (Yamamoto et al., 2015). PBMCs were frozen in foetal bovine serum (FBS) containing 10% dimethyl sulfoxide and stored in liquid nitrogen until analysis.
Method details
Establishment of HA expressing NKR-luc cells
We first synthesized cDNA encoding firefly luciferase and cloned it into the pMx-IRES-Puro Vector (Cell Biolabs, Inc., San Diego, CA, USA). To prepare luc-transduced mouse retroviruses, pMx-fLuc-IRES-Puro was transfected into the packaging Platinum-A cells (Cell Biolabs, Inc.). After 48 h of transfection, the culture supernatants were collected and infected into A549 cells to obtain A549-luc cells. To establish target cells that are resistant to NK cell-mediated spontaneous cytotoxicity, we co-cultured A549-luc cells with NK92mi for one week and KHYG-1 for another week. As a result, we created an NK-resistant cell line, A549-NKR-luc.
Next, A549-NKR-luc cells were electroporated using a 4D-Nucleofector System (Lonza, Basel, Switzerland) with pCMV/R H1 A/Puerto Rico/8/1934 (PR8) (GenBank: NP_040980) and pCMV/R H1 A/California/07/2009 (CA) (GenBank: FJ969540) A549-NKR-luc cells, respectively. The expression of surface HA was monitored using anti-human IgG (H + L) cross-adsorbed secondary antibody Alexa Fluor 647 (Thermo Fisher Scientific, Waltham, MA, USA). Cells with a high level of HA expression were sorted and selected with blasticidin (final concentration 10 μg/mL) to establish stable HA-expressing A549-NKR-luc cell lines. All cells were cultured at 37°C in a 5% CO2 humidified atmosphere.
Cynomolgus-sized monoclonal antibody
The pcDNA3.1-PD-1 cynoIgG1 heavy chain and the pcDNA6-PD-1 cynoIgG1 light chain vectors expressing the PD-1 cynoIgG1 heavy (GenBank: CS101552) and light chains (GenBank: AFF60184), respectively, were kindly gifted by Dr. Satoshi Nagata at the National Institutes of Biomedical Innovation, Health and Nutrition, Osaka, Japan. To generate a cynoIgG2 heavy chain constant region, a codon-optimized cynoIgG2 heavy chain constant region (GenBank: CS101554) fragment and cynoIgG1 LALA (L234A and L235A) were generated using a commercial gene synthesis service (GeneArt, Thermo Fisher Scientific, Waltham, MA, USA). The synthesized fragment was exchanged with cynoIgG1 constant regions in the pcDNA3.1-PD-1 cynoIgG heavy chain using NheI and NotI sites. Fv regions of the constructs were exchanged with FI6 heavy chain variable region (GenBank: JN234439) and light chain variable region (GenBank: JN234448) (Corti et al., 2011) fragments by the NEBuilder HiFi DNA Assembly (New England BioLabs, Ipswich, MA, USA).
FI6-cynoIgG1 and IgG2 were expressed using the Expi293 expression system (Thermo Fisher Scientific). Expi293F cells were cultured in FreeStyle serum-free medium (Invitrogen, Carlsbad, CA, USA) and adjusted to 1 × 106/mL in 100 mL FreeStyle serum-free medium. Each 100 μg of heavy chain and light chain plasmids and 100 μL 293fectin (Thermo Fisher Scientific) in 10 mL Opti-MEM (Thermo Fisher Scientific) were mixed and transfected into Expi293F cells. After 20 min, 100 mL FreeStyle serum-free medium was added, and the cells were incubated for 5dat 37°C in a 10% CO2 humidified atmosphere. The culture supernatants of the cells were collected and filtered through a 0.45-μm filter. The supernatant was concentrated using AKTA flux s (Cytiva, Tokyo, Japan) and diluted with an equal volume of 40 mM phosphate buffer (pH 7.0) (Nacalai Tesque, Kyoto, Japan). An automated protocol to purify cynoIgGs was established using HiTrap rProtein A FF columns (Cytiva) with AKTA Start (Cytiva). The solution was loaded onto a HiTrap rProtein A FF column equilibrated with 20 mM phosphate buffer. After washing with 20 mM phosphate buffer, FI6-cynoIgG was eluted with 100 mM glycine-HCl (pH 2.7) (Polysciences Inc., Warrington, PA, USA). Monitoring UV absorption at 280 nm facilitated fractionation and sample collection. After elution, the peak fractions were neutralized with 1 M Tris (pH 9.0) (Nippon Gene, Tokyo, Japan), dialyzed with phosphate-buffered saline (PBS) using Slide-A-Lyzer G2 Dialysis Cassettes (Thermo Fisher Scientific) at 4°C overnight. Finally, the antibody was concentrated at 1 mg/mL using an Amicon Ultra-3K (Thermo Fisher Scientific) (Doria-Rose et al., 2012; Li et al., 2012; Wu et al., 2010, 2011).
FcγR-effector analysis by flow cytometry
The frozen PBMCs of cynomolgus macaques were thawed and washed with RPMI1640 medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% heat-inactivated FBS (Sigma-Aldrich), 100 unit/mL penicillin, and 100 μg/mL streptomycin (Sigma-Aldrich) (R10), and treated with 50 units/mL benzonase (Merck, Darmstadt, Germany) in R10 for 1 h under standard conditions (37°C, 5% CO2, 95% humidity). After washing with R10, the PBMCs were labeled with the fluorescent dye 5,6-carboxyfluorescein diacetate succinimidyl ester (CFSE, Thermo Fisher Scientific) at final concentrations of 5 μM (Figure 2) and 100 nM (Figures 4 and 5) in PBS for 7 min under standard conditions, and the reaction was stopped by washing twice with ice-cold 6% FBS in PBS. The labeled PBMCs were further washed with PBS, centrifuged, resuspended in R10, and counted. HA-expressing-A549-NKR-luc cells were counted and incubated with FI6-cynoIgG1/LALA/2, control humanIgG1 (clone QA16A12), serum, or purified IgG from serum by rProtein A/Protein G GraviTrap (Cytiva) at a final concentration of 10 μg/mL for 15 min at room temperature. After incubation, unbound IgGs were washed out with PBS. The antibody-binding HA-expressing-A549-NKR-luc cells at 1 × 105 cells and the labeled PBMCs at 1 × 105 cells (Figure 2) or 5 × 105 cells (Figures 4 and 5) were co-cultured in 200 μL R10 for 15 min in a 96-well round-bottom plate (Corning, Glendale, AZ, USA). After co-culture, the cells were replaced in a 96-well V-bottom plate (Corning) and washed twice with PBS, followed by staining with the LIVE/DEAD Fixable Violet Dead Cell Stain Kit (Thermo Fisher Scientific) for 10 min at room temperature. After washing, the cells were fixed with 1% paraformaldehyde and analyzed using a FACSymphony A5 instrument equipped with five lasers (BD Biosciences, San Jose, CA, USA). Data were analyzed using the FlowJo software version 10.7.1 (BD Biosciences).
FcγR-effector analysis with subset markers
Cell suspensions containing binding cells were generated using the method described in section 2.4. After staining with the LIVE/DEAD Fixable Violet Dead Cell Stain Kit, FcR Blocking Reagent (Miltenyi Biotec, Bergisch Gladbach, Germany) was added and the cells were incubated for 10 min at room temperature. After washing with PBS, the cells were stained for anti-CD64-PE (10.1), anti-CD123-Cy7PE (7G3), anti-CD56-Alexa Fluor 700 (B159), anti-CD32-BV786 (FL18.26), anti-CD3-BUV615 (SP-34), anti-CD14-BUV805 (M5E2) (all from BD Biosciences), anti-CD11c-PE-Cy5 (3.9), anti-CD16-APC-Cy7 (3G8), anti-CD20-BV711 (2H7) (all from BioLegend), anti-CD159a (NKG2A)-APC (Z199) (Beckman Coulter, Brea, CA, USA), and anti-HLA-DR–TRPE (TU36) (Life Technologies, Carlsbad, CA, USA) for 15 min at room temperature. After washing with PBS, the cells were fixed with 1% paraformaldehyde and analyzed by flow cytometry.
Imaging cytometric analysis
PR8-HA-expressing A549-NKR-luc cells were bound to FI6-cynoIgG1 or IgG2 at a final concentration of 0.5 μg/mL for 15 min at room temperature. After washing, the cells were labeled with ER tracker Red (Thermo Fisher Scientific) for 15 min under standard conditions. The PBMCs of cynomolgus macaques were labeled with CFSE for 15 min at a final concentration of 2 nM. These cells were co-cultured for 45 min under standard conditions and labeled with Hoechst 33342 (Thermo Fisher Scientific) at a final concentration of 250 ng/mL for 15 min at room temperature. After washing with PBS twice, the cells were fixed with 1% paraformaldehyde, resuspended in PBS, and analyzed using ImageStreamX Mk II (Luminex, Austin, TX, USA). Data were analyzed and quantified using IDEAS software version 6.2 (Luminex). For the analysis, the cells were divided into three populations (R1, R2, and R3) according to the intensity of CFSE and area, referring to the plots of flow cytometry analysis (FCS-A versus CFSE intensity). Debris and off-focus cells were removed by expanding with maximum and mean pixel features using bright field images (Ch01) (R4), and 200 cells were randomly extracted for each population. Then, the population with a lobe feature ≥2 in the bright field (Ch01) was selected (R5). Using the define mask function, an object mask (tight option) was created with the mask as M01 and channel as Ch01, and the new symmetry 4 and circularity features were created based on this mask. Finally, symmetry 4 and circularity features were used to expand and divide the data into three populations (binds, singlet, and separate), and the frequency of binds was calculated (Figure S2A).
Measurement of antibody titers
The levels of total IgG in plasma from cynomolgus macaques were determined by enzyme-linked immunosorbent assay. To measure anti-HA-specific antibody titers against CA-HA, 96-well plates were coated with CA-HA protein solution overnight at 4°C. After blocking with 1% bovine serum albumin solution, the plates were washed and incubated with diluted plasma samples for 2 h at room temperature. To detect bound antibodies, the plates were washed and incubated for 1 h with biotin-conjugated anti-human total IgG antibody (BD Biosciences). Subsequently, the plates were washed and incubated for 1 h with HRP-conjugated streptavidin (Thermo Fisher Scientific). The optical density was measured at a wavelength of 450 nm. The antibody for the endpoint titer was determined with a cut-off value of 0.3. Plasma at six weeks in the vaccine schedule (Figure S4A) was diluted in PBS to match the endpoint titer at 52.5 and used for the FcγR-effector analysis.
Gene expression analysis
PBMCs were resuspended in R10 and added CA-HA-expressing A549-NKR-luc cells treated with the serum for 4 h. Subsequently, the classical monocytes were sorted from the PBMCs using a FACSymphony S6 (BD Biosciences) based on the gating strategy shown in Figure S3A. mRNA was isolated using an RNeasy MicroKit (QIAGEN, Hilden, Germany). cDNA was synthesized from purified mRNA using a SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific) and real-time PCR was performed on a Step One Plus (Thermo Fisher Scientific) system with THUNDERBIRD SYBR qPCR Mix (TOYOBO, Osaka, Japan) to measure CD80 and CD83 expression. cynoGAPDH was used as a housekeeping gene. The analyses for activation marker expression were performed based on 2−ΔΔcycle threshold method.
Quantification and statistical analysis
Statistical analyses and creation of graphs were performed using Prism 7.0a (GraphPad, San Diego, CA, USA) and SPICE software (version 6.0) (Roederer et al., 2011). Differences between two groups were compared using the Mann-Whitney U-test for unpaired samples or the Wilcoxon matched-pairs signed rank test for paired samples. Correlation analysis was performed using Spearman’s rank correlation coefficient. All p values less than 0.05 were considered significant and are indicated in the text as appropriate, ∗ <0.05, and ∗∗ <0.01.
Acknowledgments
We thank Prof. Yasuhiro Yasutomi of the Tsukuba Primate Research Center, NIBIOHN, for valuable discussions and support. We thank all members of the Laboratory of Immunosenescence, especially Mses. Eiko Moriishi, Mami Ikeda, Mika Yagoto, and Yuki Katayama for their excellent technical support. We also thank HAMRI Co., Ltd. and the Corporation for Production and Research of Laboratory Primates for their support with the animal experiments.
This study was supported by the Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (B) (grant number 20H03728) and the Japan Agency for Medical Research and Development (grant numbers 19 pc0101043h0001 and 21fk0108141h0702).
Author contributions
TY conceived and designed the experiments. YM, ST, and TN performed the experiments and analyzed the data. SM and YT contributed reagents, materials, and analysis tools. YM, ST, and TY created the figures and wrote the manuscript. All authors verified and discussed the data.
Declaration of interests
The authors declare no competing interests.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105085.
Supplemental information
Data and code availability
-
•
All data reported in this study are available from the lead contact upon request.
-
•
This study does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adachi Y., Tonouchi K., Nithichanon A., Kuraoka M., Watanabe A., Shinnakasu R., Asanuma H., Ainai A., Ohmi Y., Yamamoto T., et al. Exposure of an occluded hemagglutinin epitope drives selection of a class of cross-protective influenza antibodies. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-11821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ana-Sosa-Batiz F., Johnston A.P.R., Hogarth P.M., Wines B.D., Barr I., Wheatley A.K., Kent S.J. Antibody-dependent phagocytosis (ADP) responses following trivalent inactivated influenza vaccination of younger and older adults. Vaccine. 2017;35:6451–6458. doi: 10.1016/j.vaccine.2017.09.062. [DOI] [PubMed] [Google Scholar]
- Ana-Sosa-Batiz F., Vanderven H., Jegaskanda S., Johnston A., Rockman S., Laurie K., Barr I., Reading P., Lichtfuss M., Kent S.J. Influenza-specific antibody-dependent phagocytosis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0154461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangaru S., Lang S., Schotsaert M., Vanderven H.A., Zhu X., Kose N., Bombardi R., Finn J.A., Kent S.J., Gilchuk P., et al. A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell. 2019;177:1136–1152.e18. doi: 10.1016/j.cell.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangaru S., Zhang H., Gilchuk I.M., Voss T.G., Irving R.P., Gilchuk P., Matta P., Zhu X., Lang S., Nieusma T., et al. A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-04704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau C.M., Alter G. Extra-neutralizing FcR-mediated antibody functions for a universal influenza vaccine. Front. Immunol. 2019;10:440. doi: 10.3389/fimmu.2019.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., Corti D., Virgin H.W., Ravetch J.V. Fc-optimized antibodies elicit CD8 immunity to viral respiratory infection. Nature. 2020;588:485–490. doi: 10.1038/s41586-020-2838-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyoglu-Barnum S., Hutchinson G.B., Boyington J.C., Moin S.M., Gillespie R.A., Tsybovsky Y., Stephens T., Vaile J.R., Lederhofer J., Corbett K.S., et al. Glycan repositioning of influenza hemagglutinin stem facilitates the elicitation of protective cross-group antibody responses. Nat. Commun. 2020;11:791. doi: 10.1038/s41467-020-14579-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhns P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood. 2012;119:5640–5649. doi: 10.1182/blood-2012-01-380121. [DOI] [PubMed] [Google Scholar]
- Corti D., Cameroni E., Guarino B., Kallewaard N.L., Zhu Q., Lanzavecchia A. Tackling influenza with broadly neutralizing antibodies. Curr. Opin. Virol. 2017;24:60–69. doi: 10.1016/j.coviro.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Voss J., Gamblin S.J., Codoni G., Macagno A., Jarrossay D., Vachieri S.G., Pinna D., Minola A., Vanzetta F., et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- Cox F., Kwaks T., Brandenburg B., Koldijk M.H., Klaren V., Smal B., Korse H.J.W.M., Geelen E., Tettero L., Zuijdgeest D., et al. HA antibody-mediated FcγRIIIa activity is both dependent on FcR engagement and interactions between HA and sialic acids. Front. Immunol. 2016;7:399. doi: 10.3389/fimmu.2016.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley A.R., Ackerman M.E. Mind the gap: how interspecies variability in IgG and its receptors may complicate comparisons of human and non-human primate effector function. Front. Immunol. 2019;10:697. doi: 10.3389/fimmu.2019.00697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darricarrère N., Qiu Y., Kanekiyo M., Creanga A., Gillespie R.A., Moin S.M., Saleh J., Sancho J., Chou T.H., Zhou Y., et al. Broad neutralization of H1 and H3 viruses by adjuvanted influenza HA stem vaccines in nonhuman primates. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abe5449. [DOI] [PubMed] [Google Scholar]
- de Taeye S.W., Bentlage A.E.H., Mebius M.M., Meesters J.I., Lissenberg-Thunnissen S., Falck D., Sénard T., Salehi N., Wuhrer M., Schuurman J., et al. FcγR binding and ADCC activity of human IgG allotypes. Front. Immunol. 2020;11:740. doi: 10.3389/fimmu.2020.00740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo D.J., Palese P., Wilson P.C., Ravetch J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Invest. 2016;126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo D.J., Tan G.S., Palese P., Ravetch J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N.A., Louder M.K., Yang Z., O'Dell S., Nason M., Schmidt S.D., McKee K., Seaman M.S., Bailer R.T., Mascola J.R. HIV-1 neutralization coverage is improved by combining monoclonal antibodies that target independent epitopes. J. Virol. 2012;86:3393–3397. doi: 10.1128/JVI.06745-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florek K.R., Weinfurter J.T., Jegaskanda S., Brewoo J.N., Powell T.D., Young G.R., Das S.C., Hatta M., Broman K.W., Hungnes O., et al. Modified vaccinia virus Ankara encoding influenza virus hemagglutinin induces heterosubtypic immunity in macaques. J. Virol. 2014;88:13418–13428. doi: 10.1128/JVI.01219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossati G., Moots R.J., Bucknall R.C., Edwards S.W. Differential role of neutrophil Fcgamma receptor IIIB (CD16) in phagocytosis, bacterial killing, and responses to immune complexes. Arthritis Rheum. 2002;46:1351–1361. doi: 10.1002/art.10230. [DOI] [PubMed] [Google Scholar]
- He W., Chen C.J., Mullarkey C.E., Hamilton J.R., Wong C.K., Leon P.E., Uccellini M.B., Chromikova V., Henry C., Hoffman K.W., et al. Alveolar macrophages are critical for broadly-reactive antibody-mediated protection against influenza A virus in mice. Nat. Commun. 2017;8:846. doi: 10.1038/s41467-017-00928-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Tan G.S., Mullarkey C.E., Lee A.J., Lam M.M.W., Krammer F., Henry C., Wilson P.C., Ashkar A.A., Palese P., Miller M.S. Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. Proc. Natl. Acad. Sci. USA. 2016;113:11931–11936. doi: 10.1073/pnas.1609316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen H., Rajendran M., Choi A., Sjursen H., Brokstad K.A., Cox R.J., Palese P., Krammer F., Nachbagauer R. Influenza virus hemagglutinin stalk-specific antibodies in human serum are a surrogate marker for in vivo protection in a serum transfer mouse challenge model. mBio. 2017;8 doi: 10.1128/mBio.01463-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J.G., Ota T., Sok D., Pauthner M., Kulp D.W., Kalyuzhniy O., Skog P.D., Thinnes T.C., Bhullar D., Briney B., et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegaskanda S., Luke C., Hickman H.D., Sangster M.Y., Wieland-Alter W.F., McBride J.M., Yewdell J.W., Wright P.F., Treanor J., Rosenberger C.M., Subbarao K. Generation and protective ability of influenza virus-specific antibody-dependent cellular cytotoxicity in humans elicited by vaccination, natural infection, and experimental challenge. J. Infect. Dis. 2016;214:945–952. doi: 10.1093/infdis/jiw262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegaskanda S., Reading P.C., Kent S.J. Influenza-specific antibody-dependent cellular cytotoxicity: toward a universal influenza vaccine. J. Immunol. 2014;193:469–475. doi: 10.4049/jimmunol.1400432. [DOI] [PubMed] [Google Scholar]
- Jegaskanda S., Weinfurter J.T., Friedrich T.C., Kent S.J. Antibody-dependent cellular cytotoxicity is associated with control of pandemic H1N1 influenza virus infection of macaques. J. Virol. 2013;87:5512–5522. doi: 10.1128/JVI.03030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennewein M.F., Alter G. The immunoregulatory roles of antibody glycosylation. Trends Immunol. 2017;38:358–372. doi: 10.1016/j.it.2017.02.004. [DOI] [PubMed] [Google Scholar]
- Kanekiyo M., Joyce M.G., Gillespie R.A., Gallagher J.R., Andrews S.F., Yassine H.M., Wheatley A.K., Fisher B.E., Ambrozak D.R., Creanga A., et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 2019;20:362–372. doi: 10.1038/s41590-018-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., O'Dell S., Wilson R., Wu X., Schmidt S.D., Hogerkorp C.M., Louder M.K., Longo N.S., Poulsen C., Guenaga J., et al. HIV-1 neutralizing antibodies display dual recognition of the primary and coreceptor binding sites and preferential binding to fully cleaved envelope glycoproteins. J. Virol. 2012;86:11231–11241. doi: 10.1128/JVI.01543-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund J., Winter G., Jones P.T., Pound J.D., Tanaka T., Walker M.R., Artymiuk P.J., Arata Y., Burton D.R., Jefferis R., et al. Human Fc gamma RI and Fc gamma RII interact with distinct but overlapping sites on human IgG. J. Immunol. 1991;147:2657–2662. [PubMed] [Google Scholar]
- Maamary J., Wang T.T., Tan G.S., Palese P., Ravetch J.V. Increasing the breadth and potency of response to the seasonal influenza virus vaccine by immune complex immunization. Proc. Natl. Acad. Sci. USA. 2017;114:10172–10177. doi: 10.1073/pnas.1707950114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee R., Kanti Barman P., Kumar Thatoi P., Tripathy R., Kumar Das B., Ravindran B. Non-classical monocytes display inflammatory features: validation in sepsis and systemic lupus erythematous. Sci. Rep. 2015;5 doi: 10.1038/srep13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullarkey C.E., Bailey M.J., Golubeva D.A., Tan G.S., Nachbagauer R., He W., Novakowski K.E., Bowdish D.M., Miller M.S., Palese P. Broadly neutralizing hemagglutinin stalk-specific antibodies induce potent phagocytosis of immune complexes by neutrophils in an fc-dependent manner. mBio. 2016;7 doi: 10.1128/mBio.01624-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M.I., Holmes E.C. The evolution of epidemic influenza. Nat. Rev. Genet. 2007;8:196–205. doi: 10.1038/nrg2053. [DOI] [PubMed] [Google Scholar]
- Parsons M.S., Lee W.S., Kristensen A.B., Amarasena T., Khoury G., Wheatley A.K., Reynaldi A., Wines B.D., Hogarth P.M., Davenport M.P., Kent S.J. Fc-dependent functions are redundant to efficacy of anti-HIV antibody PGT121 in macaques. J. Clin. Invest. 2019;129:182–191. doi: 10.1172/JCI122466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond D.D., Bajic G., Ferdman J., Suphaphiphat P., Settembre E.C., Moody M.A., Schmidt A.G., Harrison S.C. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc. Natl. Acad. Sci. USA. 2018;115:168–173. doi: 10.1073/pnas.1715471115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M., Nozzi J.L., Nason M.C. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel U.J.E., Schlegel P., Lang P. Natural killer cell mediated antibody-dependent cellular cytotoxicity in tumor immunotherapy with therapeutic antibodies. Front. Immunol. 2013;4:76. doi: 10.3389/fimmu.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Chen J., Li R., Zhang M., Wang G., Stegalkina S., Zhang L., Chen J., Cao J., Bi X., et al. A multimechanistic antibody targeting the receptor binding site potently cross-protects against influenza B viruses. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aam5752. [DOI] [PubMed] [Google Scholar]
- Simhadri V.R., Dimitrova M., Mariano J.L., Zenarruzabeitia O., Zhong W., Ozawa T., Muraguchi A., Kishi H., Eichelberger M.C., Borrego F. A human anti-M2 antibody mediates antibody-dependent cell-mediated cytotoxicity (ADCC) and cytokine secretion by resting and cytokine-preactivated natural killer (NK) cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0124677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K. Avian influenza H7N9 viruses: a rare second warning. Cell Res. 2018;28:1–2. doi: 10.1038/cr.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan G.S., Krammer F., Eggink D., Kongchanagul A., Moran T.M., Palese P. A pan-H1 anti-hemagglutinin monoclonal antibody with potent broad-spectrum efficacy in vivo. J. Virol. 2012;86:6179–6188. doi: 10.1128/JVI.00469-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa O., Gilliland G.L., Brezski R.J., Strake B., Wilkinson T., Lacy E.R., Scallon B., Teplyakov A., Malia T.J., Strohl W.R. An engineered Fc variant of an IgG eliminates all immune effector functions via structural perturbations. Methods. 2014;65:114–126. doi: 10.1016/j.ymeth.2013.06.035. [DOI] [PubMed] [Google Scholar]
- Vanderven H.A., Ana-Sosa-Batiz F., Jegaskanda S., Rockman S., Laurie K., Barr I., Chen W., Wines B., Hogarth P.M., Lambe T., et al. What lies beneath: antibody dependent natural killer cell activation by antibodies to internal influenza virus proteins. EBioMedicine. 2016;8:277–290. doi: 10.1016/j.ebiom.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderven H.A., Jegaskanda S., Wines B.D., Hogarth P.M., Carmuglia S., Rockman S., Chung A.W., Kent S.J. Antibody-dependent cellular cytotoxicity responses to seasonal influenza vaccination in older adults. J. Infect. Dis. 2017;217:12–23. doi: 10.1093/infdis/jix554. [DOI] [PubMed] [Google Scholar]
- Watanabe A., McCarthy K.R., Kuraoka M., Schmidt A.G., Adachi Y., Onodera T., Tonouchi K., Caradonna T.M., Bajic G., Song S., et al. Antibodies to a conserved influenza head interface epitope protect by an IgG subtype-dependent mechanism. Cell. 2019;177:1124–1135.e16. doi: 10.1016/j.cell.2019.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle J.R.R., Zhang R., Khurana S., King L.R., Manischewitz J., Golding H., Dormitzer P.R., Haynes B.F., Walter E.B., Moody M.A., et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Yang Z.Y., Li Y., Hogerkorp C.M., Schief W.R., Seaman M.S., Zhou T., Schmidt S.D., Wu L., Xu L., et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zhou T., Zhu J., Zhang B., Georgiev I., Wang C., Chen X., Longo N.S., Louder M., McKee K., et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Lynch R.M., Gautam R., Matus-Nicodemos R., Schmidt S.D., Boswell K.L., Darko S., Wong P., Sheng Z., Petrovas C., et al. Quality and quantity of TFH cells are critical for broad antibody development in SHIVAD8 infection. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.aab3964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this study are available from the lead contact upon request.
-
•
This study does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.