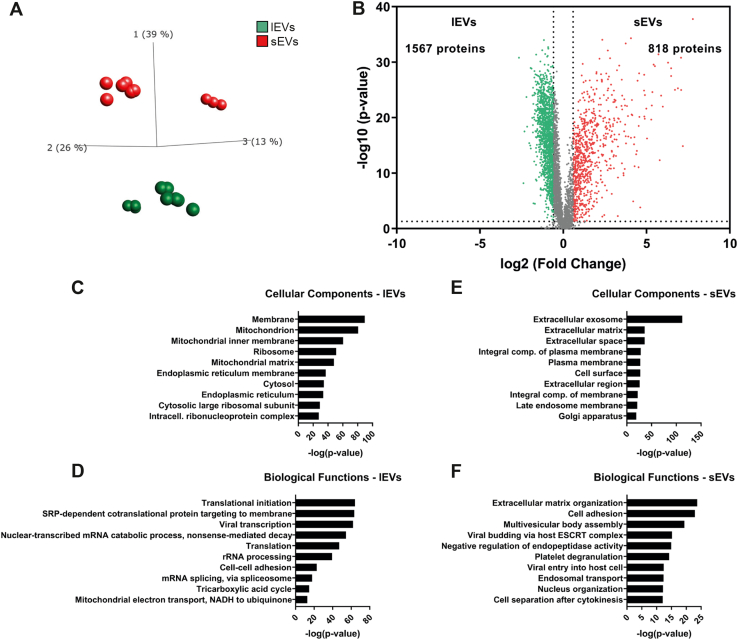
Fig. 4.
The proteomes of lEVs and sEVs are substantially dissimilar. Quantitative proteomics (tandem mass tag) was used to determine the differences in the proteomes of sEVs and lEVs. Three biological replicates (45 μg protein/sample) were used from three different cell lines resulting in N = 9. A, principal component analysis illustrating the relationship between sEVs and lEVs derived from the three cell lines. B, volcano plot of the proteomes of sEVs and lEVs identified 818 and 1567 proteins, respectively, significantly enriched more than 1.5 fold change. Dotted lines indicate cutoffs; 1.3 on the y-axis (corresponding to p < 0.05) and 0.585 on the x-axis (corresponding to fold change >1.5). C–F, Database for Annotation, Visualization and Integrated Discovery (DAVID) was used to determine the most enriched cellular compartments (C and E) and biological processes (D and F) associated with proteins significantly enriched in lEVs (C and D) and sEVs (E and F). The ten most enriched terms (based on p-value) in each category are displayed. lEV, large extracellular vesicle; sEV, small extracellular vesicle.