Fig. 1.
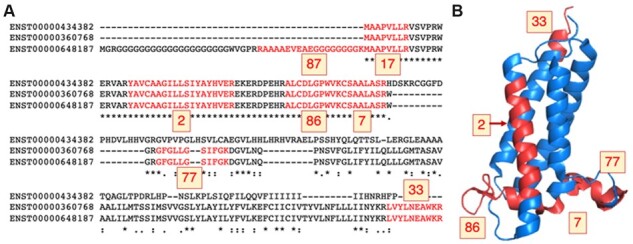
Peptides mapping to isoforms from gene VKORC1L1. (A) The alignment between the three protein sequence unique isoforms of gene VKORC1L1 (Vitamin K epoxide reductase complex subunit 1-like protein 1) with the peptides detected in the five large-scale proteomics analyses in red. The number of PSM detected for each peptide is highlighted in yellow boxes. The protein coded by transcript ENST00000648187 was chosen as the main experimental isoform because 309 PSM map to this isoform (against just 189 and 222 PSM for the other two isoforms). (B) VKORC1L1 peptides mapped to the structure of human VKOR (PDB: 6WVH, Burley et al., 2017). Peptides are marked in red on the structure and the number of PSM detected is in yellow boxes. The protein is membrane-bound and no peptides were found in membrane-spanning regions, except for peptide YAVCAAGIIISIYAYHVER (2 PSM). The N-terminal peptides (87 and 17 PSM) do not map to the structure; this region is likely to be disordered. Peptide mapping was carried out using HHPRED (Gabler et al., 2020). The image was generated with PyMol
