Figure 1.
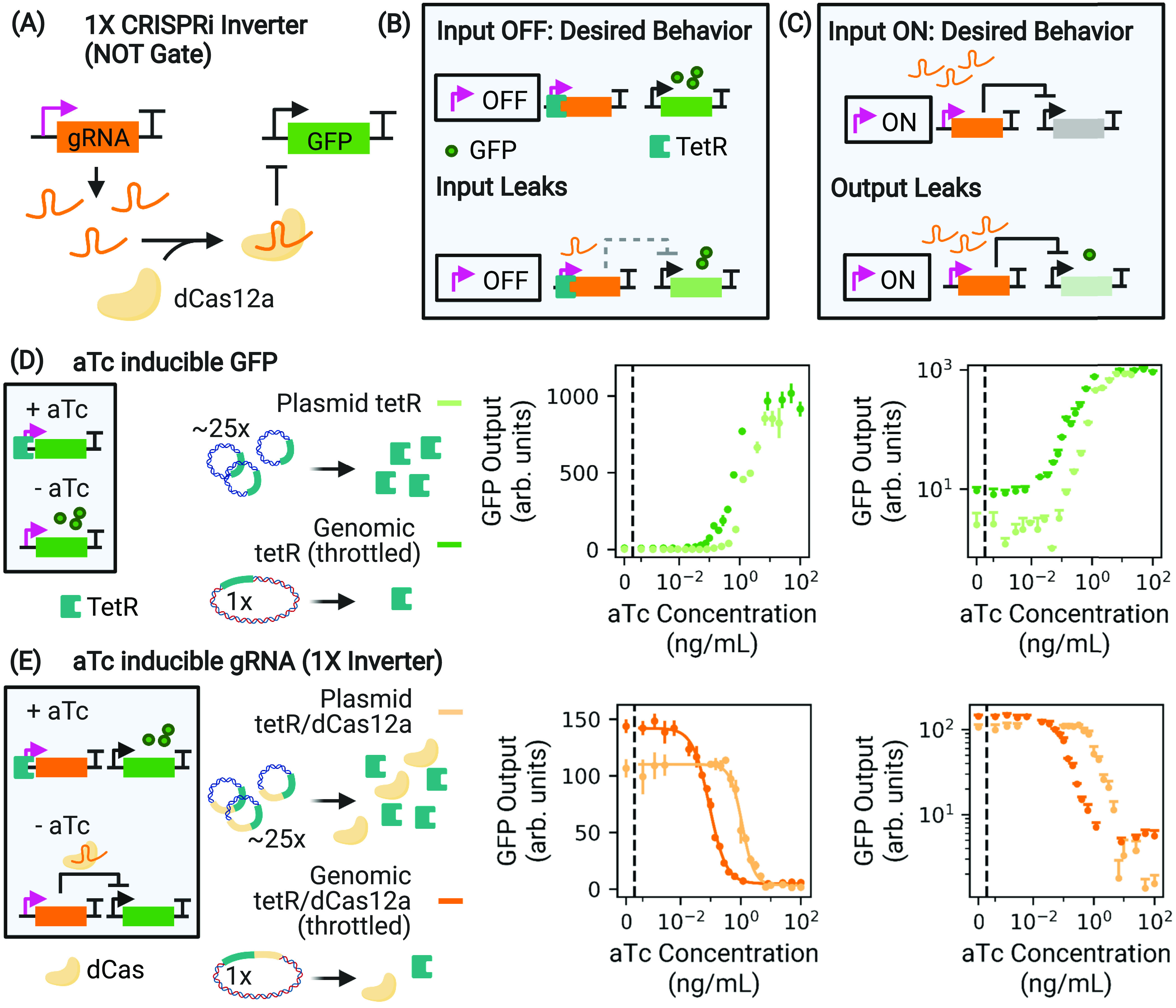
Leaks in dCas-based transcriptional circuits. (A) A CRISPRi-based NOT gate drives the production of a gRNA that programs dCas to bind to and repress expression from the target promoter, here inhibiting GFP production. (B) If the input module is an inducible sensor, any basal expression allows the unwanted production of a few gRNA that can efficiently repress the output (input leak). (C) Downstream applications can be hindered by incomplete repression by dCas (output leak). (D) We throttle tetR availability by expressing it in the genome (dark green), which causes leaky pTet expression at low aTc concentration compared to 20- to 30-fold plasmid (p15A origin, light green) expression. When used as an input promoter in an inverter, such a leaky pTet causes input leak. (E) We throttle both tetR and dCas availability, now for the 1× inverter. Throttling dCas decreases the sensitivity to leaked gRNA at low aTc concentration, increasing the overall dynamic range (dark orange) with respect to high copy plasmid expression of dCas (light orange). However, this decreases the absolute off level of GFP expression, as is evident in log space. Throttling the availability of TetR and dCas increases the leak of transcripts that they repress (gRNA and GFP mRNA, respectively), facilitating study of how these impacts can be mitigated. The curves depicted in (D) and (E) were taken during exponential growth. In linear space, the displayed error bars are ±1 standard deviation from threefold biological replicates.
