Figure 2.
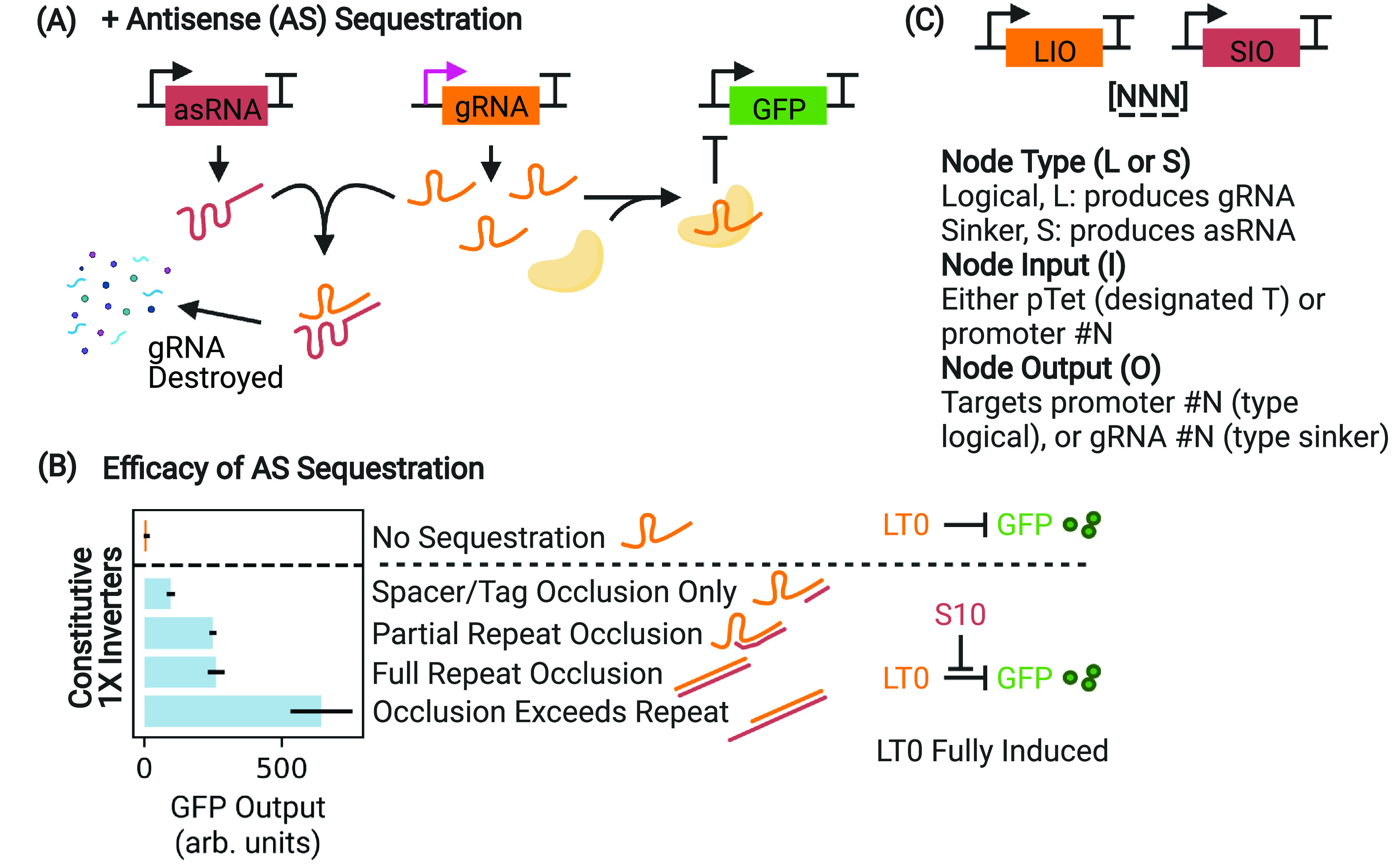
Efficacy of gRNA sequestration measured via interference with a 1× inverter. (A) By soaking up and destroying leaked gRNA transcripts using a matching asRNA sequence, the upstream circuit leak that limits the circuit dynamic range can be nullified. (B) Full expression of a 1× inverter (orange, corresponding to high aTc concentration in Figure 1D) produces cells that are white, as GFP expression is suppressed by dCas binding. Occluding portions of the gRNA (occluding only the spacer/tag, partial or full occlusion of the repeat, and occlusion that exceeds the repeat sequence, light blue) results in a demonstrable difference in sequestration efficacy as a function of interference with the function of CRISPRi, which increases GFP output. Occlusion of the complete gRNA sequence, exceeding the full length of the repeat, results in the most effective sequestration. Ultimately, partial repeat occlusion is used in all subsequent experiments in order to minimize potential nonorthogonality with asRNAs intended to target different gRNAs. Differences in GFP output are measured during exponential growth. For clarity, the HFQ recruitment tag on the asRNA is not depicted. (C) Nodes are notated with a three-character system designating the node type (logic or sinker), the promoter number, and the output number (either CRISPRi target or asRNA tag). This is useful for specifying the node order as circuits get larger and more complex. In this work, GFP is always driven by promoter 0. A pTet-driven node input is designated “T”.
