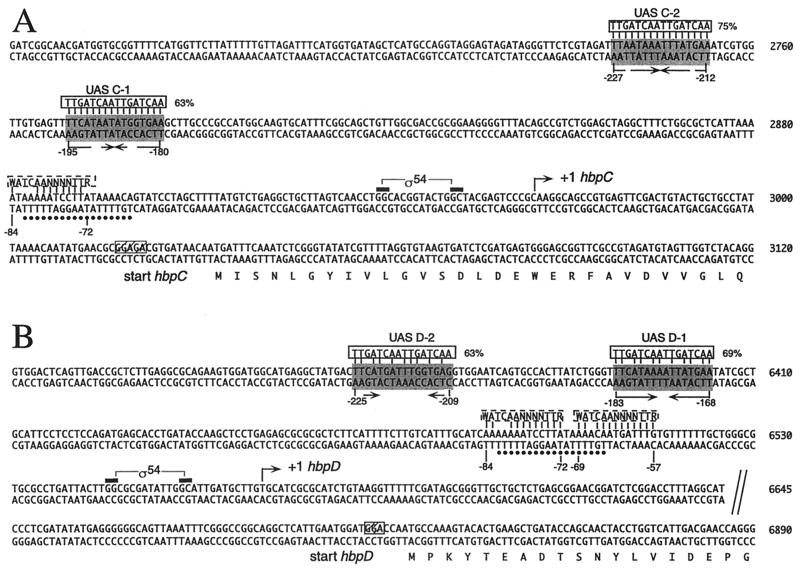
FIG. 5.
Sequences of the DNA regions upstream of the hbpC (A) and the hbpD (B) genes. The base pair numbering corresponds to that of the GenBank entry under accession no. U73900. Bent arrows, transcriptional start sites (+1) for the hbpC and hbpD genes and the direction of transcription; black bars, −24 and −12 elements in ς54-dependent promoters PhbpC and PhbpD; shaded boxes, putative UASs for HbpR binding (arrows underneath, palindromic structures within these DNA regions). Alignments with the consensus sequence for XylR/DmpR-type UASs (5′-TTGATCAATTGATCAA-3′; solid boxes) as proposed by Pérez-Martín and de Lorenzo (36) are shown at the top of putative UASs with numbers indicating the percentages of identity between the two DNA motifs. Dashed boxes, alignments of DNA regions similar to the consensus IHF binding site (5′-WATCAANNNNTTR-3′, where W = A or T, R = A or G, and N = any nucleotide) (15); dots, 18-bp-long DNA stretch overlapping the putative IHF sites and in common to both the hbpC and the hbpD promoters. The numbers below putative UASs and IHF sites indicate the relative positions with respect to the transcriptional start site. The N-terminal parts of HbpC and HbpD are indicated below the corresponding sequences. Hatched boxes, putative ribosome binding sites.