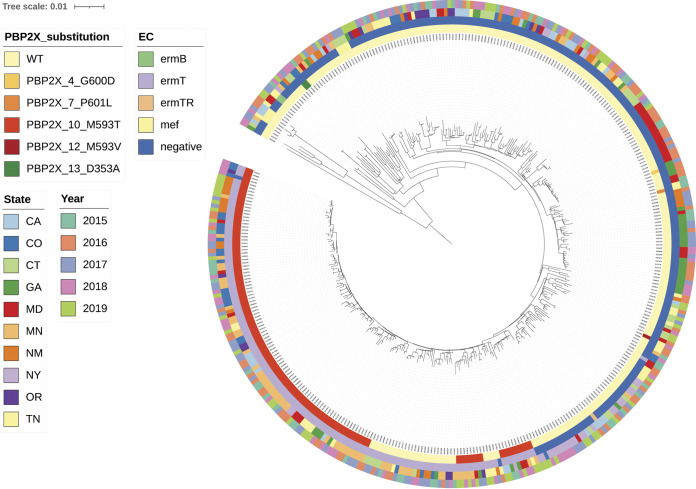
FIG 2.
Core phylogeny of emm4 isolates (all 390 recovered in 2015 to 2019 are characterized). All isolates are within clonal complex CC39. Legends for this and all other phylogenies depicted employ same scheme as follows: Innermost circle is color coded for PBP2x substitution, the second circle from inside is color coded for resistance determinants for erythromycin and clindamycin (both constitutive and inducible), the third circle from inside is depicting state, and the fourth circle from inside is depicting year of isolation. Figure depicts well established emm4/PBP2x10 sublineage recovered in multiple states and years and is the principal component of a single major branch within the overall invasive emm4 phylogeny. The emm4/ermT-positive/PBP2x1 isolates are situated on the same main branch, potentially indicating that the pbp2x10 missense mutation originated within an emm4/ermT background.