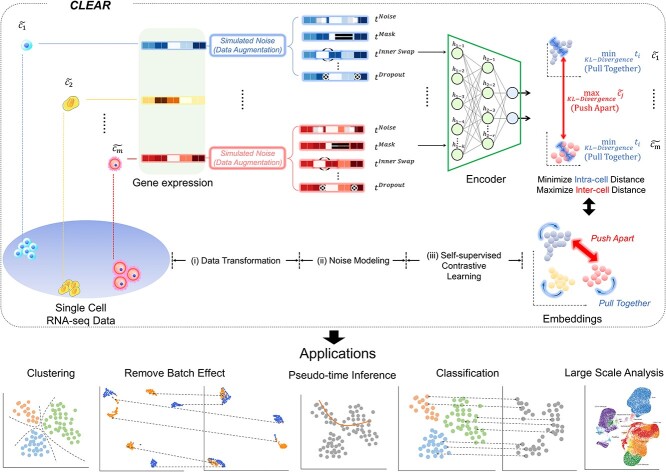
Figure 1.
Overview of the proposed framework, CLEAR. The proposed method is based on self-supervised contrastive learning. For the gene expression profile of each cell, we distort the data slightly by adding noise to the raw data, which mimics the technical noise in the biological experiments. When training the deep encoder model, we force the model to produce similar low-dimension representations for the raw data and the corresponding distorted profile while distant representations for cells of different types. Intuitively, the deep learning model learns to pull together the representations of similar cells while pushing apart different cells. By considering noise during training, CLEAR can produce effective representations while eliminating technical noise for the scRNA-seq profiles. Such representations have a broad range of applications, including clustering and classification, dropout event and batch effect correction, pseudo-time inference. CLEAR is also scalable to million-scale datasets without any overhead.