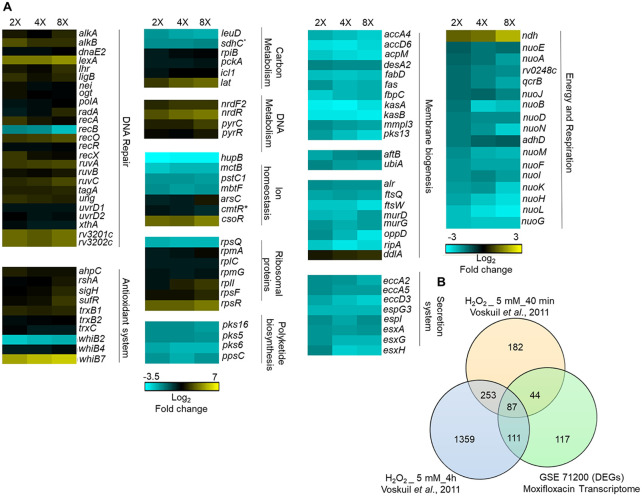
FIG 3.
Whole-genome transcriptome profiling of M. tuberculosis treated with moxifloxacin. (A) Heat map showing gene expression changes due to 16-h treatment of M. tuberculosis with moxifloxacin. DEGs exhibited a 2-fold change across all three treatment conditions (2×, 4×, and 8× MIC of moxifloxacin; 1× MIC = 0.4 μM); the color code for the fold change is at the bottom of the second column (yellow, upregulated genes; turquoise, downregulated genes). Genes are grouped according to function. For genes belonging to bioenergetics processes, the color code for the fold change is at the bottom of the fourth column. *, sdhC and cmtR are deregulated in two treatment conditions (2× and 4×). (B) Venn diagram showing transcriptome overlap between moxifloxacin-mediated (green circle) and H2O2-mediated stress for M. tuberculosis (44); DEGs obtained with treatment with 5 mM H2O2 for 40 min and 5 mM H2O2 for 4 h are shown in beige and blue circles, respectively.