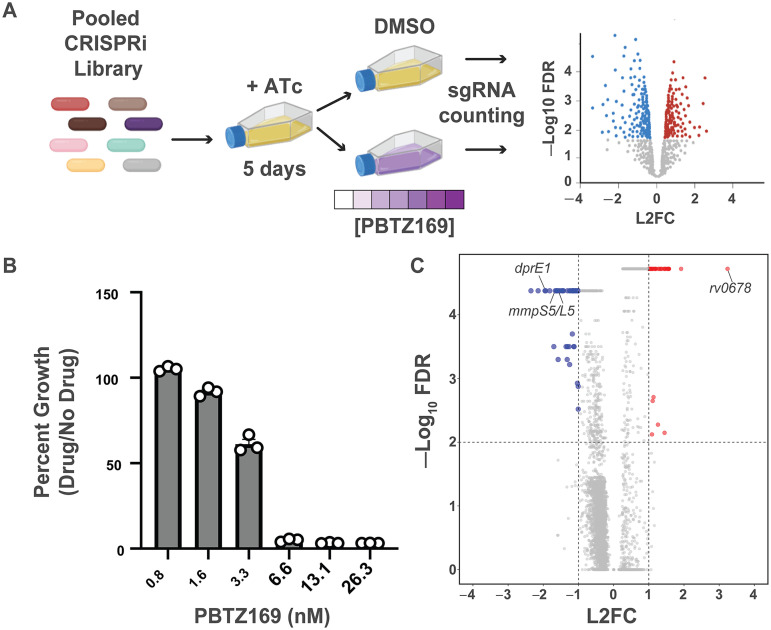
FIG 1.
CRISPRi chemical-genetic profiling of PBTZ169. (A) Chemical-genetic profiling workflow. The CRISPRi library contains 96,700 sgRNAs targeting 4,052/4,125 genes in the H37Rv genome. The library was predepleted for 5 days in the presence of anhydrotetracycline (ATc) prior to treatment with DMSO or various concentrations of PBTZ169 spanning the MIC. After 14 days of library outgrowth, genomic DNA was isolated from all cultures with detectable outgrowth. The sgRNA-encoding region was subjected to deep sequencing for control cultures as well as the 0.8 nM and 1.6 nM PBTZ169 cultures. Hit genes were called by MAGeCK (32). (B) Relative growth of PBTZ169-treated cultures. After 14 days of outgrowth, growth was calculated relative to the DMSO control. Individual replicates are shown. (C) Volcano plot summarizing the log2-fold change (L2FC) value and –log10 of the false discovery rate (FDR) for each gene in the PBTZ169 screen (1.6 nM). Key hit genes are highlighted.