Figure 4.
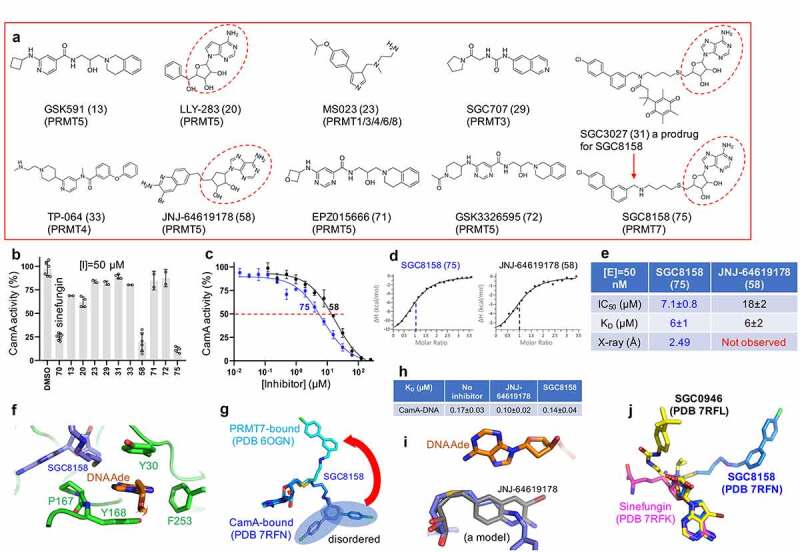
CamA inhibition by PRMT inhibitors. (a) Chemical structures of known PRMT inhibitors examined. The numerical number in parenthesis refers to our laboratory code. (b) Relative inhibition of CamA activity at a single inhibitor concentration of 50 μM. (c) The IC50 measurements by varying inhibitor concentrations of compounds SGC8158 and JNJ-64619178. (d) ITC measurement of dissociation constants and stoichiometry (vertical red dash line) of the two compounds to CamA. (e) Summary of the IC50 and KD values of inhibition of methylation of CamA by inhibitors SGC8158 and JNJ-64619178. (f) Structure of CamA-DNA in the presence of SGC8158, which becomes part of the binding site for the DNA target adenine. (g) Superimposition of SGC8158 with CamA-bound (blue) and PRMT7-bound (cyan). (h) The ITC dissociation constants of CamA-DNA were measured in the absence and presence of inhibitors (see Figure S4). (i) A model of JNJ-64619178 (grey) superimposed onto SGC8158 (blue), sitting next to target DNA adenine (orange). (j) Superimposition of CamA-bound sinefungin (magenta), SGC0946 (yellow) and SGC8158 (blue).
