ABSTRACT
Antifungal resistance to Candida pathogens increases morbidity and mortality of immunosuppressive patients, an emerging crisis worldwide. Understanding the Candida prevalence and antifungal susceptibility pattern is necessary to control and treat candidiasis. We aimed to systematically analyse the susceptibility profiles of Candida species published in the last ten years (December 2011 to December 2021) from mainland China. The studies were collected from PubMed, Google Scholar, and Science Direct search engines. Out of 89 included studies, a total of 44,716 Candida isolates were collected, mainly comprising C. albicans (49.36%), C. tropicalis (21.89%), C. parapsilosis (13.92%), and C. glabrata (11.37%). The lowest susceptibility was detected for azole group; fluconazole susceptibilities against C. parapsilosis, C. albicans, C. glabrata, C. tropicalis, C. guilliermondii, C. pelliculosa, and C. auris were 93.25%, 91.6%, 79.4%, 77.95%, 76%, 50%, and 0% respectively. Amphotericin B and anidulafungin were the most susceptible drugs for all Candida species. Resistance to azole was mainly linked with mutations in ERG11, ERG3, ERG4, MRR1–2, MSH-2, and PDR-1 genes. Mutation in FKS-1 and FKS-2 in C. auris and C. glabrata causing resistance to echinocandins was stated in two studies. Gaps in the studies’ characteristics were detected, such as 79.77%, 47.19 %, 26.97%, 7.86%, and 4.49% studies did not mention the mortality rates, age, gender, breakpoint reference guidelines, and fungal identification method, respectively. The current study demonstrates the overall antifungal susceptibility pattern of Candida species, gaps in surveillance studies and risk-reduction strategies that could be supportive in candidiasis therapy and for the researchers in their future studies.
KEYWORDS: Candida, antifungal susceptibility pattern, candidiasis, systematic review, china
Introduction
Antimicrobial resistance is a worldwide public health concern and is particularly worrisome regarding fungal infections. The fewer antifungals’ drugs for invasive infections and the emergence of multidrug-resistant (MDR) fungal pathogens have been associated with increased mortality. Candida species are the most common opportunistic pathogens that cause severe infections in the immunosuppressive host. The Candida species can cause superficial, vaginal, and oral mucosa infections and invasive candidiasis, such as deep tissue infections and bloodstream infections [1]. Candida albicans is the leading cause of candidiasis around the world. Besides this, the other common non-C. albicans species such as C. parapsilosis, C. glabrata, and C. tropicalis have emerged as health concerns over the past few decades [2]. In addition, the emergence and high prevalence of antifungal drugs resistant (AFR) C. albicans and MDR non-C. albicans species such as C. auris, have caused great concern for health care officials across the globe [1,3,4]. Phylogenetic analysis of C. auris shows five major clades that cluster geographically and are renowned for various single nucleotide polymorphisms [5,6]. Therefore, it is imperative to fully comprehend and monitor the trend of local epidemiology, and antifungal drug susceptibility to take smart decisions in the treatment of candidiasis [7].
The threat due to MDR C. auris further increases in the current COVID-19 epidemic as the ratio of hospitalized patients increases, providing a vulnerable environment for nosocomial infections. A recent study reported a mortality rate of 67.85% for coronavirus-associated C. auris infections [8]. Timely diagnosis and proper antifungal treatment are required to improve clinical outcomes and manage candidiasis [7,9]. However, laboratory-based technique like Candida culture is still inefficient for sensitive and rapid diagnosis of candidiasis. Therefore, the physicians most often prescribed empirical antifungal drugs [10,11]. The selection of empirical antifungal drugs mainly depends on epidemiological antifungal sensitivity data, which differs for every geographic region [3,7]. Hence, locally and country-wide antifungal surveillance data are required to select antifungal drugs accurately.
The molecular epidemiology of AFR Candida species is relatively less studied than the resistance mechanisms of bacteria [12]. The AFR Candida species needs to be addressed because of its high risk to human health as it ranked fourth amongst hospital-acquired and bloodstream pathogens [13]. In mainland China, the China Hospital Invasive Mycosis Surveillance Network (CHIF-NET) is an excellent project reporting antifungal susceptibility and epidemiology trends since 2009, functional in 30 out of 33 provinces (http://chifnet.com/login.cshtml). C. albicans is highly prevalent in China, but the proportion of emerging non-C. albicans species are increasing (reported >67% in 2020), mainly due to their AFR ability [14]. On this ground, several studies individually reported the epidemiology of AFR Candida species from various regions of China. However, there is no comprehensive systematic acquisition data on the antifungal susceptibility pattern of Candida from China. Therefore, in the current study, we planned to analyse the overall landscape of the distribution and antifungal susceptibility pattern of Candida from the published literature in China, which has not been documented. The outcomes of the present study will highlight the gaps in surveillance studies and provide recommendations for researchers [62]. Furthermore, the current study will provide directions for health care officials and prescribers about the resistance magnitude of Candida species to different antifungal agents, allowing them to devise strategies to combat and manage AFR in the region.
Methods
Literature search
The original research articles about Candida species in mainland China were collected from NCBI PubMed, Google Scholar, and Science Direct search engines. The preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines and checklists were followed for article selection (Figure 1) [15]. The studies were searched by using keywords like; (“Candida” AND “Antifungal resistance” AND/OR “Antifungal susceptibility” AND China); (“Candidemia” AND “Antifungal resistance” AND/OR “Antifungal susceptibility” AND China) and (“Candidiasis” AND “Antifungal resistance” AND/OR “Antifungal susceptibility” AND China). Additionally, the bibliographies of selected research articles were thoroughly reviewed to access all the studies in the domain.
Figure 1.
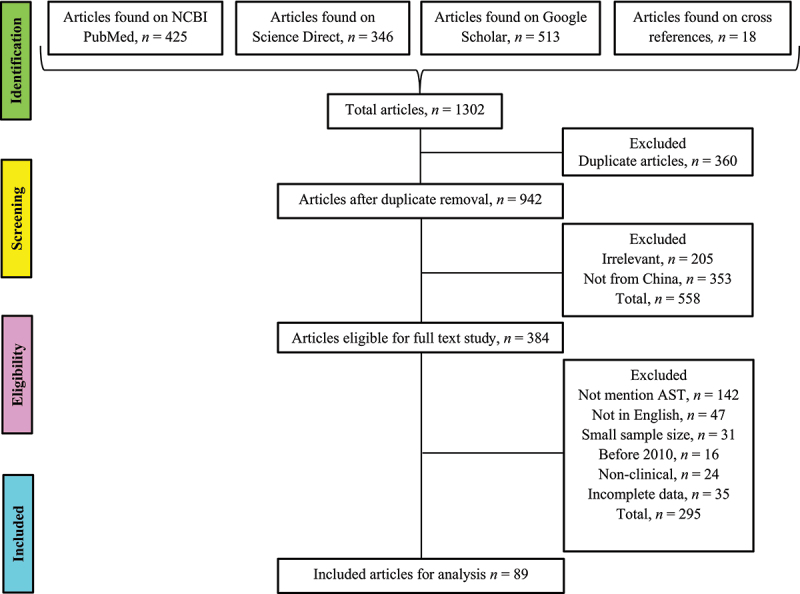
Studies identification and selection based on PRISMA guidelines.
Articles selections
All the articles found on the Databases were downloaded, and duplicates were removed. Initially, the title and abstract were reviewed to select only relevant studies from mainland China. Further article selections were performed by three researchers (HB, MS, and BH, independently) based on the full-text review of each article following the inclusion and exclusion criteria.
Inclusion criteria
The included articles had at least ten Candida species isolated from clinical specimens and mentioned the total number of isolates. The articles had data on antifungal susceptibility tests (AST), written in English and published from December 2011 till December 2021 in mainland China.
Exclusion criteria
All the articles were excluded, with isolates size less than ten, isolates not from human origin, did not perform the susceptibility tests or incomplete data, articles that did not use the standard authenticated protocols for AST, and review articles.
Data extraction
The three researchers, HB, MS, and MNK, separately collect the data from each article into a pre-prepared Excel sheet (2016). The collected data were about publication year, location of the study, sampling duration, inpatient or outpatient, gender, age, infection types, species type, total number of isolates, identification method, AST method, breakpoint reference guidelines, tested drugs, susceptibility profile, mortality rate, the molecular mechanism of AFR, and molecular phylogeny of the Candida isolates. The duration of the samples was determined by the year in which the sampling concluded. All the patients aged less than 18 years were considered paediatric. Only the susceptible or wild-type isolates percentages were counted, and the intermediate or susceptible dose-dependent were not considered.
Statistical analysis
The number of occurrences and percentages for each variable were counted in Excel sheet. The antifungal susceptibility pattern for each Candida species against every antifungal agent in the form of median susceptible or median wild type (MS/WT) with a 95 % confidence interval (CI) was determined to estimate a consistent ratio for combined data. The student’s t-test or the Wilcoxon signed-rank test was performed to measure the statistical significance of the antifungal susceptibility data against each drug. The P-values smaller than 0.05 were considered significant, calculated for two-tailed with a Gaussian approximation. For C. albicans, C. tropicalis, C. glabrata, C. parapsilosis, and C. krusei, due to isolation from different infection types, subgrouping M.S/WT, with 95% CI were also calculated. For each species, three subgroups were compiled i.e. isolates from all publications, invasive candidiasis and bloodstream infection (IC/BSI), and vulvovaginal and oral candidiasis (VVC/OC). Additionally, subgroup analysis was performed for different susceptibility testing methods i-e., BMD, ATB fungus 3, Sensititre YeastOne kit, and agar diffusion methods. The statistical analysis and graph designing were performed using the GraphPad Prism v8.0.2 software. Three researchers performed the calculation separately to negotiate any possible bias.
Results
Studies characteristics
Eighty-nine articles were included for systematic analysis containing 44,716 Candida isolates. High proportion of articles was published in 2019 and 2020 (13 in each), while for the duration of sampling, 40 studies were reported from 2010 to 2015 (supplementary file; figure S1). The maximum studies were reported from east China (n = 43), followed by north China (n = 33). Among the subregions, the highest number of articles were from Beijing (n = 20) and Shanghai (n = 18), while we did not find any study from two subregions of southwest China, i-e., Guizhou (province) and Tibet (autonomous region), and one province (Qinghai) of the northwest region (Supplementary file: figure S2). The number of articles stated the data about patient demography and AST are presented in Table 1. The median age with 95% CI from the ages mentioned articles (n = 47) was 57.45 years (53–62.5). The all-causes mortality rate was mentioned in 18 studies in which C. albicans associated median mortality was 27.3%, interquartile range (IQR) (13.60–38.30%). The median mortality rates (IQR) of Candida pathogens in included literature are stated in supplementary file (figure S3). Molecular typing of Candida isolates was performed only in 16 (17.98%) articles. The three different genotyping methods, i-e multilocus sequence typing (MLST), random amplified polymorphic DNA (RAPD) analysis, and microsatellite markers typing, were used in these studies. The detail about the molecular typing methods for the genotyping of Candida species is presented in Table 2.
Table 1.
Characteristics of articles include in current systematic analysis (n = 89)
| Characteristics | Frequency (%) | References |
|---|---|---|
| Species | ||
| C. albicans | 65 (73.03%) | (14, 26, 27, 37-42, 44, 47-54, 56-60, 67-108) |
| C. tropicalis | 47 (52.81%) | (14, 37-43, 47-49, 51, 52, 54, 57-60, 67-76, 93, 94, 97-102, 104, 107-117) |
| C. glabrata | 43 (48.31%) | (14, 17, 37-40, 42-44, 47-52, 56-59, 67-76, 96-102, 104-106, 108, 118-120) |
| C. parapsilosis | 37 (41.57%) | (5, 14, 37-41, 43, 48, 49, 51-58, 60, 71-76, 93-95, 97-102, 108, 118, 121) |
| C. krusei | 16 (17.98%) | (14, 38, 43, 44, 56, 59, 97, 99-101, 104-106, 108, 122, 123) |
| C. guilliermondii | 6 (6.74%) | (14, 97, 98, 108, 124, 125) |
| C. pelliculosa | 3 (3.37%) | (98, 102, 108) |
| C. lusitaniae | 3 (3.37%) | (14, 97, 108) |
| C. auris | 2 (2.25%) | (16, 24) |
| C. haemulonii | 1 (1.12%) | (126) |
| C. africana | 1 (1.12%) | (103) |
| Patient type | ||
| Inpatients | 35 (39.32%) | (16, 24, 26, 37-41, 47-54, 57-60, 68, 71, 72, 76, 89, 93, 102, 104, 105, 107, 111, 113, 115, 116, 118, 121) |
| Outpatients | 3 (3.37%) | (70, 80, 96) |
| Both | 12 (13.48%) | (14, 41, 55, 69, 94, 97, 98, 108, 110, 119, 120, 124) |
| NM | 39 (43.82%) | NA |
| Age group | ||
| Adult | 42 (47.19%) | (5, 24, 26, 37, 39, 40, 42, 44, 49, 51-54, 56, 57, 59, 60, 67, 69-72, 75, 78-80, 84, 85, 90, 91, 93, 95, 96, 100, 103, 106, 107, 115, 118-120, 124) |
| Adult/ Pediatrics | 18 (20.22%) | (14, 17, 38, 47, 48, 50, 55, 58, 76, 94, 97-99, 101, 102, 108, 110, 121) |
| Age group NM | 29 (32.58%) | NA |
| Gender | ||
| Female | 19 (21.34%) | (26, 44, 69, 70, 75, 77-80, 85, 89-91, 95, 96, 100, 103, 106) |
| Male/Female | 46 (51.68%) | (5, 14, 17, 24, 37-40, 42, 47-60, 67, 71, 72, 76, 87, 88, 93, 94, 97, 99, 102, 104, 107, 108, 110, 113, 115, 116, 118-121, 124) |
| NM | 24 (26.97%) | NA |
| Infection types | ||
| Invasive Candidiasis | 27 (30.33%) | (5, 14, 37, 41, 43, 47, 49, 50, 59, 60, 68, 73, 74, 94, 97-99, 102, 105, 108, 114, 115, 117, 120, 123-125) |
| Bloodstream infection | 15 (16.85%) | (17, 39, 40, 48, 51-55, 57, 58, 71, 72, 76, 93) |
| Vulvovaginal candidiasis | 20 (22.47%) | (26, 44, 69, 70, 75, 77-80, 83-85, 89-91, 95, 96, 100, 103, 106) |
| Oral candidiasis | 5 (5.61%) | (42, 56, 67, 104, 107) |
| Multiple infections | 8 (8.99%) | (16, 86, 87, 109-111, 116, 121) |
| UTI | 1 (1.12%) | (113) |
| Not Mentioned. | 13 (14.61%) | NA |
| Phenotypic identification methods | ||
| CHROMagar media | 41 (46.06%) | (26, 37, 42, 44, 47, 50, 55, 56, 59, 67-70, 72, 75-83, 85-88, 91, 95, 96, 98, 99, 101-103, 105, 106, 109, 111, 112, 123) |
| MALDI-TOF MS | 29 (32.58%) | (5, 16, 24, 38, 40, 43, 47, 48, 50, 57, 59, 67, 68, 80, 94, 97, 99, 102, 108, 110, 111, 113, 114, 116, 120, 121, 123, 124, 126) |
| API 20C AUX | 21 (23.59%) | (26, 37, 41, 42, 55, 56, 70, 75, 77, 78, 83, 85, 87, 88, 95, 98-101, 105, 122) |
| VITEK 2 COMPACT | 17 (19.10%) | (39, 41, 51-54, 58, 60, 67, 69, 72, 73, 76, 82, 96, 99, 106) |
| BD BACTEC™ FX | 5 (5.61%) | (39, 40, 52, 53, 93) |
| VITEK MS system | 2 (2.24%) | (14, 16) |
| Microscopy | 2 (2.25%) | (85, 103) |
| DL-96A ID/AST | 2 (2.25%) | (71, 118) |
| yeast identification kit | 1 (1.12%) | (109) |
| ATB ID32C strips | 1 (1.12%) | (27) |
| Brilliance Candida agar | 1 (1.12%) | (41) |
| NM | 4 (4.49%) | NA |
| Molecular identification methods | ||
| ITS sequencings | 32 (35.95%) | (5, 14, 16, 24, 38, 41-43, 55, 67, 72-74, 79, 84, 85, 90, 92, 94, 97, 99, 102, 104, 107, 108, 110, 115, 120, 123-126) |
| D1/D2 analysis | 6 (6.74%) | (24, 37, 41, 73, 84, 110) |
| Molecular beacon assay | 1 (1.12%) | (96) |
| Susceptibility testing method | ||
| Broth microdilution | 35 (39.32%) | (14, 17, 26, 56, 60, 67, 72-74, 77, 79-83, 85, 86, 89-92, 97, 99-101, 106, 109, 112-116, 120, 122, 123) |
| ATB FUNGUS 3 kit | 24 (26.97%) | (27, 37, 39, 40, 47, 49-54, 57-59, 68-70, 78, 87, 88, 93, 96, 105, 119) |
| Sensititre Yeast-1 kit | 16 (17.98%) | (5, 16, 24, 38, 42, 43, 48, 94, 102, 110, 111, 117, 120, 121, 124, 126) |
| Agar Diffusion method | 12 (13.48%) | (41, 44, 75, 76, 95, 98, 103, 104, 107, 108, 110, 118) |
| ETEST method | 2 (2.25%) | (37, 55) |
| kit of Autobio | 1 (1.12%) | (71) |
| Neo-Sensitabs | 1 (1.12%) | (84) |
| Breakpoint reference guidelines | ||
| CLSI | 82 (92.13%) | (5, 14, 16, 17, 24, 26, 27, 37-43, 47-52, 55-57, 59, 60, 67-70, 72-75, 77-83, 85-126) |
| EUCAST | 3 (3.37%) | (40, 52, 93) |
| NM | 7 (7.86%) | NA |
Footnote: The sum of percentages is not equal to 100 because some studies stated more than one variable, and we counted each study separately with each variable.
NM= Not mentioned, NA= Not applicable, UTI= urinary tract infection, ITS=internal transcribed spacer, CLSI= clinical laboratory and standard institutes, EUCAST= European committee of antimicrobial sensitivity testing
Table 2.
The molecular typing of Candida species mentioned in the included studies
| Species/ Method | Outcomes/Detail | References |
| C. albicans | ||
| MLST | DSTs: 79, 435, 1867, 365, 1395, 254, 443, 365, and CC 69 |
(26, 27) |
| Microsatellite markers | CAI and CP6 | (37, 72) |
| RAPD analysis | Primer: 5’-ACGGCCGACC-3’ and 5’-CCAGATGCAC-3’ |
(78, 87) |
| C. tropicalis | ||
| MLST | DSTs; 225, 639, 329, 615, 506, 508, 519, 169, 345, 399, 300, 546, 376, 321, 322, 323, 324, 325, 114, 169, 326, 327, 328, 331, 332, 333, 334, 335, 336, 337, 338, 339, 340, 341, 164, 341, 342, 343, 344, 346, 347, 348, 349, 99, 350, 351, 352, 353, 354, 355, 356, 181, 615, 852, 984, 996, 991,482. 901, 330, 980, 337, 522, 990, 982, 998, 723, 983, 997, 977, 730, 489, 993, 403, 184, 139, 833, 889, 999, 995, 992, 986, 434, 981, 994, 394, 994, 437, 978, 525, 985, 923, 979, 986, 987, 988, 1000, 1001, 1002, |
(72, 111, 113, 115, 116) |
| Microsatellite markers | ctm1, ctm3, ctm24 | (117) |
| C. glabrata | ||
| MLST | STs: ST7, ST10, ST22, ST45, ST55, ST3, |
(17, 72, 120) |
| Microsatellite markers | GLM5, RPM2, GLM4, ERG3, MTI, GLM6, | (17, 120) |
| C. parapsilosis | ||
| Microsatellite markers | CAI, CP6, B5, CP1, CP4, | (72, 121) |
| C. krusei | ||
| Microsatellite markers | 33 markers were selected; Cakr001–Cakr033 | (123) |
| C. guilliermondii | ||
| Microsatellite markers | sc15, sc32, sc72 | (125) |
Footnote: DSTs; Diploid sequence types, ST; Sequence types, MLST; multilocus sequence type. RAPD; Randomly amplified polymorphic DNA.
Prevalence of Candida species
C.albicans accounted for 49.36% of all Candida isolates reported in 65 articles. Among the non-C. albicans species, C. tropicalis (21.89%) was the most prevalent specie, followed by C. parapsilosis (13.92%) and C. glabrata (11.37%). C. africana (0.03%) was the most rarely reported Candida species. The percentages and number of all Candida species reported in the current study are presented in Figure 2(a). Broadly, mainland China is divided into seven regions, among which differences in the distribution of Candida species in various regions were observed. The highest number of isolates were reported from north China (23.07%), followed by east (15.20%), south (13.95%), northeast (11.12%), southwest (9.11%), central (8.69%), and northwest region (8.20%), while 10.64% of the isolates were from multiple locations. C. albicans and C. tropicalis were highly reported in the north (23.61% and 24.67%), followed by east (17.22%, and 14.08%) and south China (16.67%, and 13.57%). C. parapsilosis proportion was highest in the north (17.63%), followed by the east (15.63%) and northeast region (14.52%). C. glabrata was highest in the north (23.48%), northeast (13.99%), and south China (12.13%). C. krusei was high in the southwest (18.56%) while C. auris were only reported in northeast China. The complete depiction of Candida species distribution in various regions of China is presented in Figure 2(b).
Figure 2.
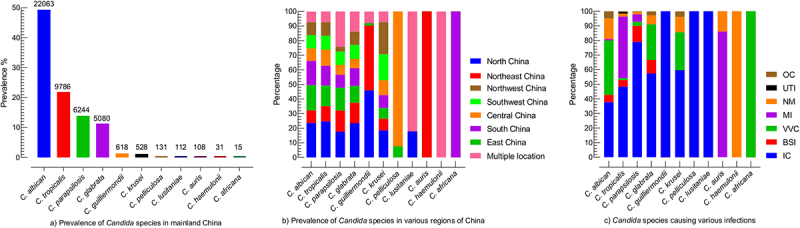
Prevalence of Candida species, (a) the total occurrence of Candida species, the numerical on the top of the bar is the number of specific Candida species, (b) the occurrence of Candida species in different regions of China, each region is represented by specific colour as shown in the box, Multiple locations mean the that studies mentioned more than one region of China, (c) Prevalence of Candida species in association with various infection type, OC; oral candidiasis, UTI; urinary tract infection, NM; not mentioned the infection type, MI; multiple infections, involved in more than one infections, VVC; vulvovaginal candidiasis, BSI; bloodstream infection, IC; invasive candidiasis.
Regarding infections type, the high proportion of Candida species was associated with invasive candidiasis (IC) (49.36%), followed by vulvovaginal candidiasis (VVC) (22.17%), bloodstream infections (BSI) (6.07%), and oral candidiasis (OC) (3.04%), while 10.31% of isolates were reported with multiple infections and for 8.09% strains the infections types were not mentioned. C. albicans were almost equally reported for IC and VVC (37.805% and 37.23% out of 22,063 isolates). C. tropicalis and C. parapsilosis were highly associated with IC (48.30%/9786 and 78.93%/6244). Similarly, C. glabrata and C. krusei were highly reported with IC (57.44% and 59.65%) followed by VVC (24.35% and 25.94%) out of 5080 and 528 isolates, respectively. Among 108 C. auris isolates, 86.11% of isolates were involved in multiple infection types, while for 13.89% of isolates, the infection types were not mentioned. The percentages of Candida species associated with various infection types are shown in Figure 2(c).
Antifungal susceptibility pattern
The median susceptibility or wild-type (M.S/WT) with a 95% confidence interval (CI) was calculated for C. albicans, C. tropicalis, C. glabrata, C. parapsilosis, C. krusei, C. guilliermondii, C. pelliculosa, C. lusitaniae, and C. auris. The C. haemulonii and C. africana were reported in one study; therefore, their MS was not determined. The susceptibility rates of all antifungal agents were statistically significant (P = < 0.05) except few; terbinafine against C. albicans was not significant (P = 0.4164), voriconazole against C. auris (P = 0.4114), clotrimazole in case of C. glabrata (P = 0.1158), and clotrimazole, ketoconazole, and miconazole against C. tropicalis were not significant (P = 0.3308, 0.0574, and 0.1065, respectively). The P- values of antifungal agents against all tested Candida isolates are mentioned in supplementary file (table S1). For the subgroup analysis based on different susceptibility testing methods, it was observed that the susceptibility rates obtained by agar diffusion methods are comparatively low than BMD, ATB fungus 3, and Sensititre YeastOne kit method almost for all antifungal agents against all tested Candida species (supplementary file; table S2)
AST of 22,063 C. albicans were performed against 14 antifungal drugs, in which high proportions of isolates were tested against fluconazole, itraconazole, voriconazole, and amphotericin B. The highest susceptibility was observed for polyenes, followed by echinocandins and 5-flucytosine. The azole class was comparatively less susceptible as the MS (95%CI) for miconazole, ketoconazole, and fluconazole were 52.95% (40–97.60), 85.3% (32.5–99.60), and 91.6% (85.85–95), respectively. In the IC/BSI group, C. albicans were tested against nine antifungal drugs, with susceptibility rates greater than 95% for all tested drugs. The micafungin and amphotericin B were the most susceptible drugs against C. albicans in VVC/OC group, having MS (95%CI) of 100% (93.80–100) and 99.4% (97.50–100), while the lowest susceptibilities were reported against miconazole (52.95% (40–97.60)) and itraconazole (79% (55.10–90.60)). AST pattern of C. albicans against all tested drugs and subgroups analysis is presented in Figure 3. AST of C. tropicalis was examined against 13 antifungal drugs, in which 100% susceptibility was reported against amphotericin B, while posaconazole (MS; 45.95, 95%CI; (29.3–88.1)) was less susceptible among all tested drugs. The susceptibility against echinocandins was in the range of 98–99%. Among the azole class, the highest MS (95%CI) was observed for voriconazole (83.96%, (73–89.4)) while fluconazole (77.95%, (65.4–83.3%)) was among the less susceptible drugs. For 5-flucytosine, variations in susceptibilities were reported between IC/BSI and VVC/OC groups, in which the MS (95% CI) were 99.8% [68–71] and 92.57% (89.7–100), respectively. The AST pattern of C. tropicalis against all antifungal drugs is depicted in Figure 4.
Figure 3.
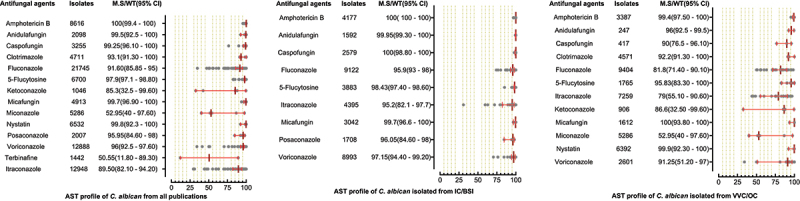
Antifungal susceptibility patterns of Candida albicans in the form of median susceptibility/wild type with 95% confidence interval.
Figure 4.
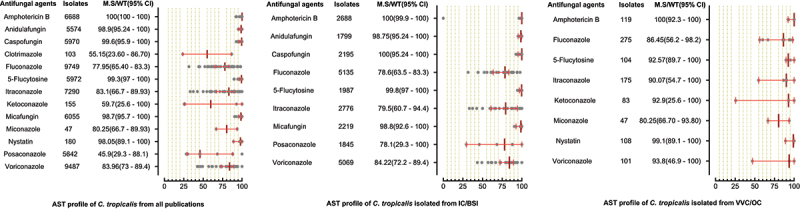
Antifungal susceptibility patterns of Candida tropicalis in the form of median susceptibility/wild type with 95% confidence interval.
A total of 6244 C. parapsilosis isolates were examined for AST and showed the highest susceptibility rates to polyenes and echinocandins compared to the azole class. The susceptibilities of C. parapsilosis against all drugs were comparatively high than other Candida species. None of the isolates was resistant to amphotericin B, while the MS (95% CI) against fluconazole was 93.25% (88.8–95.3). The susceptibilities of the VVC/OC group containing C. parapsilosis against echinocandins and azole drugs were comparatively low than isolates of the IC/BSI group (Figure 5). For C. glabrata the lowest MS (95%CI) was reported against ketoconazole (48.7% (42.1–62.1)), followed by clotrimazole (73.7% (16.6–91.9)) and fluconazole (79.4% (54.4–86.4)), while was 100% susceptible against amphotericin B. Overall, the C. glabrata of the IC/BSI group were more susceptible to antifungal drugs than VVC/OC group containing isolates. The complete antifungal susceptibility pattern of C. glabrata is presented in Figure 6. C. krusei were 100% susceptible against amphotericin B and to two drugs of echinocandin class i-e., anidulafungin and micafungin, while the lowest MS (95%CI) were observed against caspofungin (93,75% (86.70–100). Similarly, in IC/BSI group, the lowest MS (95%CI) was reported against caspofungin (91.5% (86.7–100)), while other echinocandins, 5-flucytosine, and polyene were 100% susceptible (Figure 7). Among the rare Candida species, the lowest MS (95% CI) was found against fluconazole, which was 94.6% [59] - [67], 76% (46.2–90.9), 50% (44.80–80.5), in the case of C. lusitaniae, C. guilliermondii, and C. pelliculosa respectively. None of the C. auris isolates was susceptible to fluconazole, while the highest MS (95%CI) was observed against itraconazole (96.75% (93.50–100)). The complete susceptibility pattern of C. guilliermondii, C. pelliculosa, C. lusitaniae, and C. auris is presented in Figure 8.
Figure 5.
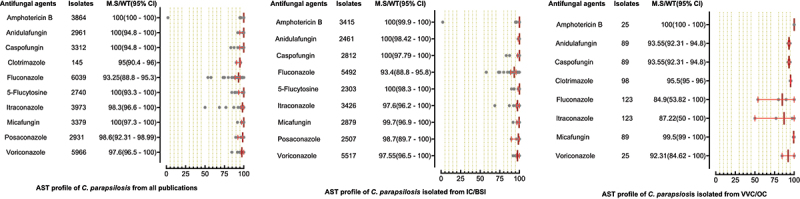
Antifungal susceptibility patterns of Candida parapsilosis in the form of median susceptibility/wild type with 95% confidence interval.
Figure 6.
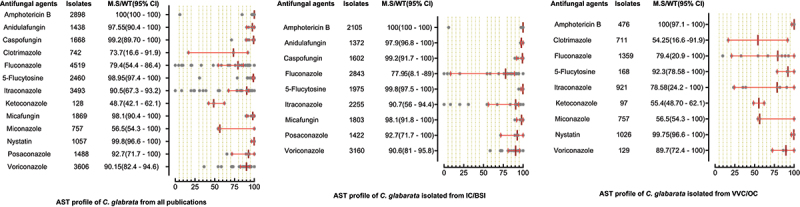
Antifungal susceptibility patterns of Candida glabrata in the form of median susceptibility/wild type with 95% confidence interval.
Figure 7.
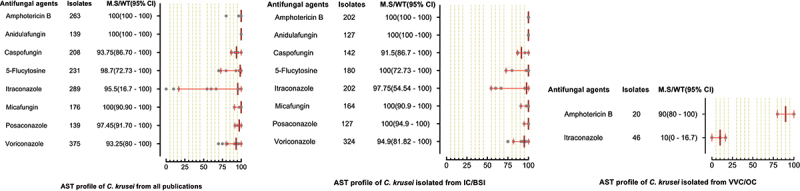
Antifungal susceptibility patterns of Candida krusei in the form of median susceptibility/wild type with 95% confidence interval.
Figure 8.
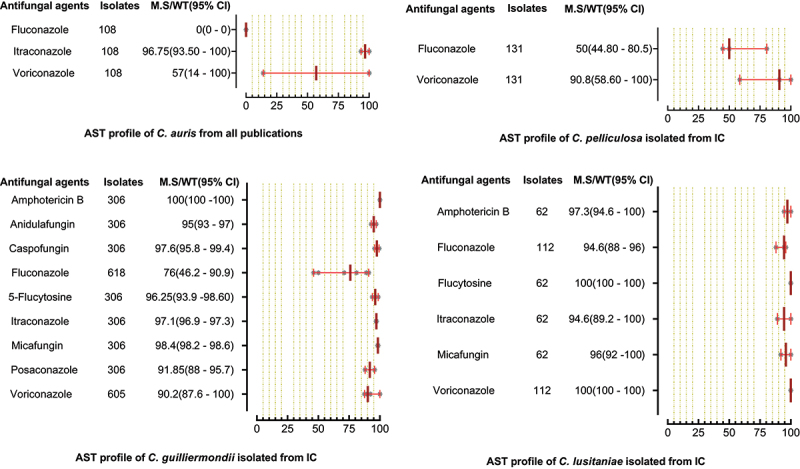
Antifungal susceptibility patterns of C. auris, C. pelliculosa, C. guilliermondii and C. lusitaniae in the form of median susceptibility/wild type with 95% confidence interval.
Molecular mechanism of antifungal resistance
Among the selected articles, 25 studies reported mutations in ergosterol biosynthesis genes lead to azole resistance. Only two studies reported echinocandins resistance due to FKS1 mutation in C. auris and FKS2 mutation in C. glabrata [90,100]. The gene mutations were detected mainly by PCR amplification and sequence analysis, except for one study that performed the whole-genome sequence of C. auris [100]. Resistance to azole due to the overexpression of regulatory genes was also detected in some studies by qRT PCR. The overall detail about the mutations in various genes, leading to amino acid substitutions and overexpression of genes causing the antifungal resistance in different Candida species from the included literature are mentioned in Table 3.
Table 3.
Molecular mechanisms of antifungal drug resistance stated in the included studies
| Mechanism | Detail | Outcome | Reference |
|---|---|---|---|
| C. albicans | |||
| ERG11 mutation |
Amino acid substitution: A114S*, Y132H*, F449Y*, Q474K*, T123I*, Y257H*, G448E*, G464S*, F72S*, G450E*, Y132F*, K143Q*, K143R*, Y257H*, D116E, P56S, V88I, V437I, E266D, V488I, D116E/D, R523G, N440S, A114V, T285A, S457P, I333T, S24G, K90R, N168S, V234G, N237D, E260G, T285A, A317T, K344E, E517V, V332L, K128T, G448R, V488I, G448R/G, G465S, D225H, S199N, F104V, L935S, E260V, N435V, G472R and D502E, D117E, E165K, E174A, V234F, G346A, A434V, and L480F |
Azole resistance | (26, 27, 70, 74, 80, 82, 83, 86, 88, 125) |
| Mrr2 mutation |
Amino acid substitution: L144V*, T145A*, V451A*, S466L*, A468G*, S480P*, H31Y, T83A, H149Y, S165N, T386I and D404N |
Azole resistance | (85) |
| ERG4 mutation |
Silent mutation: CA133 and C435T |
Azole resistance | (86) |
| ERG3 mutation |
Amino acid substitution: A18P/A, R365G/R, W219C and R352H Silent mutations: T342G, T435C, C441T, T1047C |
Azole resistance | (26, 89) |
| Efg1 mutation |
Amino acid substitution: V86I Silent mutation: A150T, A165C, G210A, G267A, G279A, A285T, A744C, A786G, T954A, C1071T, A1055C, A1317G |
Azole resistance | (89) |
| Cap1 mutation |
Amino acid substitution: A390T, S381N, P311S, G481E |
Azole resistance | (90) |
| Mrr1 mutation |
Amino acid substitution: R557K*, K844E*, F1032L*, S1037L*, N937K, E1020Q, T917M and T923I |
Azole resistance | (90) |
| Overexpression | ADH1, CDR1, CDR2, MDR1, FLU1, ERG11, TAC1, Cap1, MDR1, Mrr1 | Azole resistance | (26, 27, 77, 81, 85, 90, 106) |
| C. glabrata | |||
| ERG 11 mutation |
Mutations: T1328C, T1394C, G1487A, A1583G Amino acid substitution= no |
Fluconazole resistance | (119) |
| MSH2 mutation |
Polymorphism: V239L, E456D, R636 M |
Azole resistance | (17) |
| PDR1 mutation |
Amino acid substitution: G348C*, N764D*, E259G, E555D, N764D, D876N, S98L, R250K, G389V, R293G, S942F, L139I, E818G, Y336H, I378T, L669F, A848V, P76S, P143T, D243N, I91V, L732S, P927S |
Azole resistance | (17, 119) |
| FKS2 mutation | S663F and S663P mutation in FKS2 HS1 | Echinocandins resistance | (17) |
| Overexpression | CDR1, CDR2, | Azole resistance | (119) |
| C. tropicalis | |||
| ERG 11 mutation |
Amino acid substitution: Y132F*, S154C*, A114S*, Y132H*, V125A, Y257H, and G464S |
Azole resistance | (68, 94, 111, 112, 114) |
| Overexpression | Erg11, CDR1, MDR1 | Azole resistance | (114) |
| C. krusei | |||
| ERG 11 mutation |
Codon substitution: T939C, T642C, A756T |
Azole resistance | (122) |
| Overexpression | Erg11 | Azole resistance | (122) |
| C. auris | |||
| ERG 11 mutation | VF125AL and I74L mutation | Azole resistance | (16) |
| FKS1 mutation | S639F mutation | Echinocandins resistance | (16) |
| C. guilliermondii | |||
| ERG 11 mutation |
Non-synonymous mutations: W37C, P518R, P430Q, D492N, Y41F, L328T, S346T, V410M, S420T, F39L, N485K, Y132F, S346T, G16S, M332I, R247K, G459S, G459K, K143R, Q469K |
Azole resistance | (125) |
| C. haemulonii | |||
| ERG 11 mutation |
Amino acid substitution: Y132H* |
Azole resistance | (94) |
*= Validated: mean all the amino acids found only in resistant strains and not in the susceptible isolates, and the authors discussed their azole resistance validity.
Discussion
The current study systematically analysed the distribution and AST pattern of Candida isolates from mainland China to provide reference points for upcoming studies. Among the selected articles, C. albicans was found in a high proportion (49.36% of all the isolates). C. albicans is a worldwide highly prevalent opportunistic pathogen [103]. Among the non-C. albicans species, C. tropicalis (21.89%) was the most prevalent, followed by C. parapsilosis (13.92%) and C. glabrata (11.37%). This drift contrasts with many European countries and North America, where the C. glabrata is more prevalent among the non-C. albicans isolates [104]. However, a similar trend of Candida species prevalence resembling our study was also reported in India, Nigeria, and Cameroon [105]. Among the infection types, a high proportion of C. albicans was associated with VVC. This is mainly due to the colonization of C. albicans in the human vagina, and getting the opportunity causes vaginal infection [106]. The invasive candidiasis and bloodstream infections are also reported in association with C. albicans and non-C. albicans species, The three main factors i.e. misuse of broad-spectrum antibiotics, cytotoxic chemotherapy-induced mucositis, and iatrogenic immunosuppression, are the causes of the high proportion of invasive candidiasis [107]. The rare Candida species were not explicitly observed with candidemia. However, the C. guilliermondii, C. pelliculosa, C. lusitaniae, and C. auris were reported to cause invasive candidiasis. This is mainly due to their MDR properties which lead to treatment failure and a longer duration of invasive candidiasis [108].
The overall C. auris clades reported in China belong to South Asian and South African clades [100,101]. The discovery of C. auris in multiple clades is mainly due to the increasing business exchange and global travelling in recent years, as reported previously in the United States [109]. In the included articles, only two studies performed the MLST of C. albicans, revealed that most of the strains belong to novel DSTs, which indicates that genetic diversity is largely unknown. However, most of the DSTs known strains belong to clades 1 and 18, suggesting their nosocomial dissemination [16,17]. Interestingly, the worldwide distributed clade of C. albicans is clade 2, which is not identified in China [17,110]. It might be due to the highly distinct global distribution of C. albicans clades. Furthermore, it is also reported that different body sites are associated with different clades, like Candida of superficial infections mostly belongs to clade 1 and BSI from clade 4 [17]. Further studies regarding molecular epidemiology targeting different body sites and populations are required.
The summary of the antifungal susceptibility profile showed that testing of azole, echinocandins, and polyenes is standard in China. In most Candida species, low susceptibility was observed for the azole group. Like in C. albicans the lowest susceptibility rates were observed for miconazole, ketoconazole, and fluconazole; 52.95%, 85.3%, and 91.60%, respectively. Similar trends of lowest susceptibility trend of azoles were also observed for non-C. albicans species. For C. tropicalis, the fluconazole and itraconazole susceptibility rates were 77.95% and 83.1%, respectively. Likewise, the fluconazole susceptibility rates for C. glabrata and C. parapsilosis were 79.4% and 93.25%, respectively. The C. glabrata and C. tropicalis are considered to have low susceptibility against azole worldwide. In the United States, Australia, and several European countries, 85% to 94% of C. glabrata isolates were reported susceptible to fluconazole [1,111,112]. In the United States, the C. tropicalis isolates were >97% susceptible to fluconazole [113]. For Latin America, Asia -Pacific regions, and Chile, nearly 90% of Candida isolates were azole susceptible [1,14,111]. The azole susceptibility of the current study is lower compared to other regions of the world and from tested polyenes and echinocandins drugs in the present study. This might be due to the high use of azole drugs in China, leading to molecular alteration of ergosterol biosynthetic pathways [114,115]. The reasons for their often prescriptions are that they are economical, available for oral administration, and reveal less toxicity [116]. The C. auris was reported in only two studies, which showed 100% resistance to fluconazole and 57% susceptibility to voriconazole. The highest susceptibility to amphotericin B was observed for all Candida species. It is mainly due to its less prescription as it is costly and causes severe renal toxicity [117]. The rapid emergence of echinocandins resistance has been observed worldwide, owing to their high use. In the United States, > 10% of C. glabrata isolates showed resistance to echinocandins [3]. In the current study, the echinocandins resistance is relatively low (0.8–2.5%) against C. glabrata. Their accurate prescriptions and usage must be maintained as the chances of acquiring resistance to echinocandins due to mutations in hotspot regions of FKS are very high [118]. The variation in susceptibility pattern of certain Candida species against azole drugs depends on various factors. The susceptibility reported in the early years was high compared to detected later in 2020 and 2021, which indicates that the susceptibility decreases due to misuse of azole drugs. For example, a study reported 85% fluconazole susceptibility in 2013, while in 2020, 48% susceptibility was reported against C. glabrata [18–21]. Likewise, a study for C. parapsilosis in 2012 reported 98.6% susceptibility, while 76.93% susceptibility was reported in 2020 against itraconazole [22,23]. Likewise, a study in 2014 reported 100% itraconazole susceptibility rate against C. krusei, while in 2016, a study reported 16.7% susceptibility rate [24,80]. Also, we reported that the median susceptibility values obtained by agar diffusion methods are lower than other methods in most cases (Supplementary table S2). Our finding contradicts earlier studies in 2002 and 2007, as they reported the same results for agar diffusion and other methods [45,119]. Further comparative studies need to evaluate the results of antimicrobial susceptibility methods based on the molecular known susceptible and resistant magnitudes. Additionally, many other factors like geography, study population, usage of azole drugs, and isolates obtained from various body sites cause variation in the anti-azole susceptibility pattern among various studies [46, 120].
Many samples were isolated from the long-term ICU, and hospitalized patients indicated that nosocomial disease is an important predisposing factor for candidiasis [20,25–31,101]. Besides this, the co-morbidities like hypoproteinemia, cardiovascular disease, diabetes, respiratory dysfunction, renal failure, cancer, HIV, and thrombocytopenia are reported to be significant for candidiasis development [21,27,29,30,32,33,94]. Apart from this, catheterization, mechanical ventilation, elderly patients, empirical antifungal therapy, and surgeries are secondary risk factors [21,26,27,29–37,94,101]. The risk factors reported in the present study are comparable to the findings of other studies [105,61,121,122]. The health care workers need special attention to hand washing or decontamination and provide an aseptic environment to minimize the horizontal transmission of candidiasis in the hospital.
The overall proportion of molecular studies regarding the genotyping and resistant mechanisms was unsatisfactory. The data obtained from molecular analysis give in-depth knowledge about the epidemiological origin and source of infections. Based on this, the health official eradicates the pathogens from their origins and inhibits their emergence and transmissions. Moreover, the molecular AFR study is essential to understand the intrinsic and acquired resistance mechanism, which may help to prevent the resistance and to design alternative and novel therapeutic agents [63,123]. To provide new insights, further molecular studies with a precise mode of action through genotyping and resistant mechanisms are required to understand pathogenicity and resistant magnitude.
Many gaps in the surveillance studies were noted, i.e. we did not find any study according to our inclusion criteria from Qinghai and Tibet Autonomous Region. The mortality rate was not mentioned in 79.77% of the studies. Nine studies did not mention the mortality rate concerning specific Candida pathogen. Similarly, 43.82% of the studies did not mention the patient types, as the inpatients are more vulnerable to candidiasis. Moreover, 47.19 %, 26.97%, 7.86%, and 4.49% of the studies did not mention the age, gender, breakpoint reference guidelines, and identification method, respectively. More importantly, such gaps in the studies make the data suspicious and difficult to analyse for practical applications. We recommend that the researchers fulfill these gaps in their future studies.
The current study focuses on the antifungal susceptibility pattern of Candida species from mainland China; however, its implication is worldwide. It is well known that resistant species are rarely limited to specific locations; any area with high drug resistant strains can act as a reservoir, from which the resistant species can be transmitted to other parts of the globe via humans, water, agricultural products, and animals [64,124]. Moreover, the current study is a point of reference for subsequent studies, as we identified the gaps in surveillance studies that need to be addressed in future studies. The information provided here may help for the development of treatment recommendations in the future that may need to be regionally tailored. Additionally, this study would dictate the overall AST profiles of Candida species, which will help the policymakers and health care officials to eradicate and exert a curative effect of antifungal agents dealing with candidiasis in mainland China.
Limitations
Most of the studies included in this systematic report were from Beijing (n = 20) and Shanghai (n = 18) regions; there is a risk of selection bias. However, Beijing is political, and Shanghai is the economic hub of China, and people from all around the country are directly or indirectly connected with these two cities. There is also a chance of bias due to the not mentioned patient type in numerous studies; usually, the Candida species isolated from inpatients are less susceptible than outpatient [65,125]. In addition, it would be preferable to include studies that contain at least 30 isolates to ensure high-quality data accuracy [66,126]. However, due to the fewer existing studies, inclusion criteria regarding the isolates in each study had set at ten to cover high number of studies. Furthermore, data acquired through various methods from diverse patient groups were assembled in the present study. However, many studies used microdilution methods and followed CLSI guidelines; the degree of variation should be marginal.
Conclusion
This study summarizes the antifungal susceptibility pattern of Candida species from mainland China and found that azole had the lowest, while amphotericin B and anidulafungin had the highest susceptibility rates among the tested antifungal drugs. The clinician can select their empirical antifungal therapy based on the outcomes of the current study. We noted substantial gaps in the surveillance studies, like no studies were found in Qinghai and Tibet regions among the included articles. Also, the number of articles for some Candida species was insufficient to calculate their susceptibility pattern. Furthermore, fewer studies performed genotyping and molecular analysis of antifungal resistance. Therefore, continuous molecular surveillance studies by researchers focusing on the mentioned gaps are of paramount importance in combating candidiasis. Along with that, precautionary measures from health staff following the guidelines of health care policymakers are necessary to halt the nosocomial dissemination and antifungal drug resistance.
Supplementary Material
Acknowledgments
The authors are thankful to the second affiliated hospital of Shantou University Medical College for supporting this study
Funding Statement
This study is funded by the Second Affiliated Hospital of Shantou University Medical College to Hazrat Bilal and Yuebin Zeng.
Contributors
Study idea and plan: HB and YZ, Attainment of data: HB, MS and MNK, Analysis and interpretation of data: HB, MNK, BH, and XY, Drafting of the manuscript: HB, RI and RUK., Critical revision of the manuscript for important intellectual content: YZ, BH, and MS., Administrative, technical, material support, and institutional study supervision: YZ. All the authors read and approved the final version of the manuscript.
Data availability statement
Data supporting our study can be provided by demand from the corresponding author (email: zeng_yb@163.com) or the first author (email: bilal.microbiologist@yahoo.com).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplemental material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2022.2123325.
References
- [1].Pfaller MA, Diekema DJ, Turnidge JD, et al. Twenty years of the SENTRY antifungal surveillance program: results for Candida Species from 1997-2016. Open Forum Infect Dis. 2019;6(Suppl 1):S79–S94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Talapko J, Juzbašić M, Matijević T, et al. Candida albicans-The virulence factors and clinical manifestations of infection. J Fungi (Basel). 2021;7(2): DOI: 10.3390/jof7020079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Perlin DS, Rautemaa-Richardson R, Alastruey-Izquierdo A.. The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect Dis. 2017;17(12):e383–e92. [DOI] [PubMed] [Google Scholar]
- [4].Jeffery-Smith A, Taori SK, Schelenz S, et al. Candida auris: a Review of the Literature. Clin Microbiol Rev. 2018;31(1): DOI: 10.1128/CMR.00029-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Guo J, Zhang M, Qiao D, et al. Prevalence and antifungal susceptibility of Candida parapsilosis species complex in Eastern China: a 15-year retrospective study by ECIFIG. Front Microbiol. 2021;12:644000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chow NA, de Groot T, Badali H, et al. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis. 2019;25(9):1780–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pappas PG, Kauffman CA, Andes DR, et al. Clinical practice guideline for the management of Candidiasis: 2016 update by the infectious diseases society of America. Clin Infect Dis. 2016;62(4):e1–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Vaseghi N, Sharifisooraki J, Khodadadi H, et al. Global prevalence and subgroup analyses of coronavirus disease (COVID-19) associated Candida auris infections (CACa): a systematic review and meta-analysis. Mycoses. 2022;65(7):683–703. DOI: 10.1111/myc.13471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kollef M, Micek S, Hampton N, et al. Septic shock attributed to Candida infection: importance of empiric therapy and source control. Clin Infect Dis. 2012;54(12):1739–1746. [DOI] [PubMed] [Google Scholar]
- [10].Chindamporn A, Chakrabarti A, Li R, et al. Survey of laboratory practices for diagnosis of fungal infection in seven Asian countries: an Asia Fungal Working Group (AFWG) initiative. Med Mycol. 2018;56(4):416–425. DOI: 10.1093/mmy/myx066 [DOI] [PubMed] [Google Scholar]
- [11].Wickes BL, Wiederhold NP. Molecular diagnostics in medical mycology. Nat Commun. 2018;9(1):5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Amanati A, Badiee P, Jafarian H, et al. Impact of antifungal stewardship interventions on the susceptibility of colonized Candida species in pediatric patients with malignancy. Sci Rep. 2021;11(1):14099. DOI: 10.1038/s41598-021-93421-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kotthoff-Burrell E. Candidemia (blood infection) and other Candida infections. Am J Respir Crit Care Med. 2019;200(5):9–p10. [DOI] [PubMed] [Google Scholar]
- [14].Xiao M, Chen SC, Kong F, et al. Distribution and antifungal susceptibility of candida species causing Candidemia in China: an update from the CHIF-NET study. J Infect Dis. 2020;221(Suppl 2):S139–s47. DOI: 10.1093/infdis/jiz573 [DOI] [PubMed] [Google Scholar]
- [15].Ziegler A, König IR. Guidelines for research reports: German translation of CONSORT 2010, PRISMA and STARD. Dtsch Med Wochenschr. 2011;136(8):357–358. [DOI] [PubMed] [Google Scholar]
- [16].Wu Y, Li C, Wang Z, et al. Clonal spread and azole-resistant mechanisms of non-susceptible Candida albicans isolates from vulvovaginal candidiasis patients in three Shanghai maternity hospitals. Med Mycol. 2018;56(6):687–694. DOI: 10.1093/mmy/myx099 [DOI] [PubMed] [Google Scholar]
- [17].Hu L, Du X, Li T, et al. Genetic and phenotypic characterization of Candida albicans strains isolated from infectious disease patients in Shanghai. J Med Microbiol. 2015;64(Pt 1):74–83. DOI: 10.1099/jmm.0.080200-0 [DOI] [PubMed] [Google Scholar]
- [18].Li F, Wu L, Cao B, et al. Surveillance of the prevalence, antibiotic susceptibility, and genotypic characterization of invasive candidiasis in a teaching hospital in China between 2006 to 2011. BMC Infect Dis. 2013;13:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Song Y, Chen X, Yan Y, et al. Prevalence and antifungal susceptibility of pathogenic yeasts in China: a 10-year retrospective study in a teaching hospital. Front Microbiol. 2020;11:1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang W, Song X, Wu H, et al. Epidemiology, species distribution, and predictive factors for mortality of candidemia in adult surgical patients. BMC Infect Dis. 2020;20(1):506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zheng YJ, Xie T, Wu L, et al. Epidemiology, species distribution, and outcome of nosocomial Candida spp. bloodstream infection in Shanghai: an 11-year retrospective analysis in a tertiary care hospital. Ann Clin Microbiol Antimicrob. 2021;20(1):34. DOI: 10.1186/s12941-021-00441-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang H, Xiao M, Chen SC, et al. In vitro susceptibilities of yeast species to fluconazole and voriconazole as determined by the 2010 National China Hospital Invasive Fungal Surveillance Net (CHIF-NET) study. J Clin Microbiol. 2012;50(12):3952–3959. DOI: 10.1128/JCM.01130-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Xiao JL, Xu GC, de Hoog S, et al. Oral prevalence of Candida species in patients undergoing systemic glucocorticoid therapy and the antifungal sensitivity of the isolates. Infect Drug Resist. 2020;13:2601–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang FJ, Zhang D, Liu ZH, et al. Species distribution and in vitro antifungal susceptibility of Vulvovaginal Candida isolates in China. Chin Med J (Engl). 2016;129(10):1161–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zeng Z, Ding Y, Tian G, et al. A seven-year surveillance study of the epidemiology, antifungal susceptibility, risk factors and mortality of candidaemia among paediatric and adult inpatients in a tertiary teaching hospital in China. Antimicrob Resist Infect Control. 2020;9(1):133. DOI: 10.1186/s13756-020-00798-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yang ZH, Song YG, Li RY. A ten-year retrospective study of invasive Candidiasis in a tertiary hospital in Beijing. Biomed Environ Sci. 2021;34(10):773–788. [DOI] [PubMed] [Google Scholar]
- [27].Pu S, Niu S, Zhang C, et al. Epidemiology, antifungal susceptibilities, and risk factors for invasive candidiasis from 2011 to 2013 in a teaching hospital in southwest China. J Microbiol Immunol Infect. 2017;50(1):97–103. DOI: 10.1016/j.jmii.2015.01.005 [DOI] [PubMed] [Google Scholar]
- [28].Zeng Z, Tian G, Ding Y, et al. Epidemiology, antifungal susceptibility, risk factors and mortality of invasive candidiasis in neonates and children in a tertiary teaching hospital in Southwest China. Mycoses. 2020;63(11):1164–1174. [DOI] [PubMed] [Google Scholar]
- [29].Xiao Z, Wang Q, Zhu F, et al. Epidemiology, species distribution, antifungal susceptibility and mortality risk factors of candidemia among critically ill patients: a retrospective study from 2011 to 2017 in a teaching hospital in China. Antimicrob Resist Infect Control. 2019;8:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang W, Song X, Wu H, et al. Epidemiology, risk factors and outcomes of Candida albicans vs. non-albicans candidaemia in adult patients in Northeast China. Epidemiol Infect. 2019;147:e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li D, Xia R, Zhang Q, et al. Evaluation of candidemia in epidemiology and risk factors among cancer patients in a cancer center of China: an 8-year case-control study. BMC Infect Dis. 2017;17(1):536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Xiao G, Liao W, Zhang Y, et al. Analysis of fungal bloodstream infection in intensive care units in the Meizhou region of China: species distribution and resistance and the risk factors for patient mortality. BMC Infect Dis. 2020;20(1):599. DOI: 10.1186/s12879-020-05291-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li YY, Chen WY, Li X, et al. Asymptomatic oral yeast carriage and antifungal susceptibility profile of HIV-infected patients in Kunming, Yunnan Province of China. BMC Infect Dis. 2013;13:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liu F, Zhong L, Zhou F, et al. Clinical features, strain distribution, antifungal resistance and prognosis of patients with non-albicans Candidemia: a retrospective observational study. Infect Drug Resist. 2021;14:3233–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bai Y, Zheng Z, Liu T, et al. Epidemiological characteristics and drug resistance of Fungemia in general hospitals from 2010 to 2019. Biomed Res Int. 2021;2021:2529171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zeng ZR, Tian G, Ding YH, et al. Surveillance study of the prevalence, species distribution, antifungal susceptibility, risk factors and mortality of invasive candidiasis in a tertiary teaching hospital in Southwest China. BMC Infect Dis. 2019;19(1):939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang H, Liu N, Yin M, et al. The epidemiology, antifungal use and risk factors of death in elderly patients with candidemia: a multicentre retrospective study. BMC Infect Dis. 2014;14:609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yu SY, Zhang L, Chen S, et al. Candida isolates causing refractory or recurrent oropharyngeal candidiasis in 11 hospitals in China. Infect Drug Resist. 2019;12:865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang B, He X, Lu F, et al. Candida isolates from blood and other normally sterile Foci from ICU patients: determination of epidemiology, antifungal susceptibility profile and evaluation of associated risk factors. Front Public Health. 2021;9:779590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Luo X, Dong X, Pen Z. Distribution and drug susceptibility of Candida spp. Associated with female genital tract infection, Chongqing, China. Jundishapur J Microbiol. 2016;9(10):e19386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yang L, Su MQ, Ma YY, et al. Epidemiology, species distribution, antifungal susceptibility, and ERG11 mutations of Candida species isolated from pregnant Chinese Han women. Genet Mol Res. 2016;15(2):1–8. [DOI] [PubMed] [Google Scholar]
- [42].Lin S, Chen R, Zhu S, et al. Candidemia in adults at a tertiary hospital in China: clinical characteristics, species distribution, resistance, and outcomes. Mycopathologia. 2018;183(4):679–689. DOI: 10.1007/s11046-018-0258-5 [DOI] [PubMed] [Google Scholar]
- [43].Dong D, Li Z, Zhang L, et al. Clinical and microbiological investigation of fungemia from four hospitals in China. Mycopathologia. 2015;179(5–6):407–414. DOI: 10.1007/s11046-014-9855-0 [DOI] [PubMed] [Google Scholar]
- [44].Pfaller MA, Castanheira M, Messer SA, et al. In vitro antifungal susceptibilities of isolates of Candida spp. And Aspergillus spp. From China to nine systemically active antifungal agents: data from the SENTRY antifungal surveillance program, 2010 through 2012. Mycoses. 2015;58(4):209–214. [DOI] [PubMed] [Google Scholar]
- [45].Zhang H, Tan J, Kontoyiannis DP, et al. Screening the in vitro susceptibility of posaconazole in clinical isolates of Candida spp. And Aspergillus spp. And analyzing the sequence of ERG11 or CYP51A in non-wild-type isolates from China. Diagn Microbiol Infect Dis. 2019;95(2):166–170. DOI: 10.1016/j.diagmicrobio.2019.05.003 [DOI] [PubMed] [Google Scholar]
- [46].Liu XP, Fan SR, Peng YT, et al. Species distribution and susceptibility of Candida isolates from patient with vulvovaginal candidiasis in Southern China from 2003 to 2012. Journal de Mycologie Medicale. 2014;24(2):106–111. [DOI] [PubMed] [Google Scholar]
- [47].Ma CF, Li FQ, Shi LN, et al. Surveillance study of species distribution, antifungal susceptibility and mortality of nosocomial candidemia in a tertiary care hospital in China. BMC Infect Dis. 2013;13:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Guo H, Zhang XL, Gao LQ, et al. Alcohol dehydrogenase I expression correlates with CDR1, CDR2 and FLU1 expression in Candida albicans from patients with vulvovaginal candidiasis. Chin Med J (Engl). 2013;126(11):2098–2102. [PubMed] [Google Scholar]
- [49].Ying C, Zhang H, Tang Z, et al. Antifungal susceptibility and molecular typing of 115 Candida albicans isolates obtained from vulvovaginal candidiasis patients in 3 Shanghai maternity hospitals. Med Mycol. 2016;54(4):394–399. [DOI] [PubMed] [Google Scholar]
- [50].Yan L, Wang XD, Seyedmousavi S, et al. Antifungal susceptibility profile of Candida Albicans isolated from Vulvovaginal Candidiasis in Xinjiang Province of China. Mycopathologia. 2019;184(3):413–422. DOI: 10.1007/s11046-018-0305-2 [DOI] [PubMed] [Google Scholar]
- [51].Wang B, Huang L, Zhao J, et al. ERG11 mutations associated with azole resistance in Candida albicans isolates from vulvovaginal candidosis patients. Asian Pac J Tropical Biomedicine. 2015;5:909–914. [Google Scholar]
- [52].Shi C, Liu J, Li W, et al. Expression of fluconazole resistance-associated genes in biofilm from 23 clinical isolates of Candida albicans. Braz J Microbiol. 2019;50(1):157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ying Y, Zhao Y, Hu X, et al. In vitro fluconazole susceptibility of 1,903 clinical isolates of Candida albicans and the identification of ERG11 mutations. Microbial Drug Resist (Larchmont, NY). 2013;19(4):266–273. DOI: 10.1089/mdr.2012.0204 [DOI] [PubMed] [Google Scholar]
- [54].Liu JY, Shi C, Wang Y, et al. Mechanisms of azole resistance in Candida albicans clinical isolates from Shanghai, China. Res Microbiol. 2015;166(3):153–161. [DOI] [PubMed] [Google Scholar]
- [55].Shi XY, Yang YP, Zhang Y, et al. Molecular identification and antifungal susceptibility of 186 Candida isolates from vulvovaginal candidiasis in southern China. J Med Microbiol. 2015;64(Pt 4):390–393. DOI: 10.1099/jmm.0.000024 [DOI] [PubMed] [Google Scholar]
- [56].Feng W, Yang J, Ji Y, et al. Mrr2 mutations and upregulation are associated with increased fluconazole resistance in Candida albicans isolates from patients with vulvovaginal candidiasis. Lett Appl Microbiol. 2020;70(2):95–101. DOI: 10.1111/lam.13248 [DOI] [PubMed] [Google Scholar]
- [57].Feng W, Yang J, Xi Z, et al. Mutations and/or Overexpressions of ERG4 and ERG11 genes in clinical Azoles-Resistant isolates of Candida albicans. Microbial Drug Resist (Larchmont, NY). 2017;23(5):563–570. DOI: 10.1089/mdr.2016.0095 [DOI] [PubMed] [Google Scholar]
- [58].Xu H, Liu M, Chen Y, et al. Randomly amplified polymorphic deoxyribonucleic acid (DNA) analysis of Candida albicans isolates from clinical sources of hospital in south China. Afr J Microbiol Res. 2012;6(10): 2552–2558. [Google Scholar]
- [59].Zhang L, Yang HF, Liu YY, et al. Reduced susceptibility of Candida albicans clinical isolates to azoles and detection of mutations in the ERG11 gene. Diagn Microbiol Infect Dis. 2013;77(4):327–329. [DOI] [PubMed] [Google Scholar]
- [60].Feng W, Yang J, Xi Z, et al. Regulatory role of ERG3 and Efg1 in Azoles-Resistant strains of Candida albicans isolated from patients diagnosed with Vulvovaginal Candidiasis. Indian J Microbiol. 2019;59(4):514–524. DOI: 10.1007/s12088-019-00833-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Feng W, Yang J, Yang L, et al. Research of Mrr1, Cap1 and MDR1 in Candida albicans resistant to azole medications. Exp Ther Med. 2018;15(2):1217–1224. DOI: 10.3892/etm.2017.5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tang Y, Yu F, Huang L, et al. The changes of antifungal susceptibilities caused by the phenotypic switching of Candida species in 229 patients with vulvovaginal candidiasis. J Clin Lab Anal. 2019;33(1):e22644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Li HM, Shimizu-Imanishi Y, Tanaka R, et al. White-Opaque switching in different mating type-like locus gene types of clinical Candida albicans Isolates. Chin Med J (Engl). 2016;129(22):2725–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Yang ZT, Wu L, Liu XY, et al. Epidemiology, species distribution and outcome of nosocomial Candida spp. bloodstream infection in Shanghai. BMC Infect Dis. 2014;14:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Chen J, Hu N, Xu H, et al. Molecular epidemiology, antifungal susceptibility, and virulence evaluation of Candida isolates causing invasive infection in a tertiary care teaching hospital. Front Cell Infect Microbiol. 2021;11:721439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhu Y, Shan Y, Fan S, et al. Candida parapsilosis sensu stricto and the closely related species Candida orthopsilosis and Candida metapsilosis in vulvovaginal candidiasis. Mycopathologia. 2015;179(1–2):111–118. [DOI] [PubMed] [Google Scholar]
- [67].Zhai Y, Liu J, Zhou L, et al. Detection of Candida species in pregnant Chinese women with a molecular beacon method. J Med Microbiol. 2018;67(6):783–789. DOI: 10.1099/jmm.0.000740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Guo LN, Yu SY, Xiao M, et al. Species distribution and antifungal susceptibility of invasive Candidiasis: a 2016-2017 multicenter surveillance study in Beijing, China. Infect Drug Resist. 2020;13:2443–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Li W, Hu YA, Li FQ, et al. Distribution of yeast isolates from invasive infections and their in vitro susceptibility to antifungal agents: evidence from 299 cases in a 3-year (2010 to 2012) surveillance study. Mycopathologia. 2015;179(5–6):397–405. DOI: 10.1007/s11046-015-9858-5 [DOI] [PubMed] [Google Scholar]
- [70].Guo LN, Xiao M, Cao B, et al. Epidemiology and antifungal susceptibilities of yeast isolates causing invasive infections across urban Beijing, China. Future Microbiol. 2017;12:1075–1086. [DOI] [PubMed] [Google Scholar]
- [71].Shi Y, Zhu Y, Fan S, et al. Molecular identification and antifungal susceptibility profile of yeast from vulvovaginal candidiasis. BMC Infect Dis. 2020;20(1):287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zhang L, Zhou S, Pan A, et al. Surveillance of antifungal susceptibilities in clinical isolates of Candida species at 36 hospitals in China from 2009 to 2013. Int J Infect Dis. 2015;33:1–4. [DOI] [PubMed] [Google Scholar]
- [73].Xu H, Yu SY, Zhou ML, et al. Epidemiology and antifungal susceptibility patterns of invasive fungal infections from 2012 to 2014 in a teaching hospital in central China. Infect Drug Resist. 2019;12:3641–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Shan Y, Fan S, Liu X, et al. Prevalence of Candida albicans-closely related yeasts, Candida africana and Candida dubliniensis, in vulvovaginal candidiasis. Med Mycol. 2014;52(6):636–640. [DOI] [PubMed] [Google Scholar]
- [75].Wu J, Guo H, Yi G, et al. Prevalent drug resistance among oral yeasts from asymptomatic patients in Hainan, China. Mycopathologia. 2014;177(5–6):299–307. DOI: 10.1007/s11046-014-9747-3 [DOI] [PubMed] [Google Scholar]
- [76].Zeng X, Peng M, Liu G, et al. Strain distribution and drug susceptibility of invasive fungal infection in clinical patients with systemic internal diseases. Front Bioeng Biotechnol. 2020;8:625024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Zhang JY, Liu JH, Liu FD, et al. Vulvovaginal candidiasis: species distribution, fluconazole resistance and drug efflux pump gene overexpression. Mycoses. 2014;57(10):584–591. DOI: 10.1111/myc.12204 [DOI] [PubMed] [Google Scholar]
- [78].Wu J, Gan C, Li J, et al. Species diversity and antifungal susceptibilities of oral yeasts from patients with head and neck cancer. Infect Drug Resist. 2021;14:2279–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Xiao M, Sun ZY, Kang M, et al. Five-Year national surveillance of invasive Candidiasis: species distribution and Azole susceptibility from the China Hospital Invasive Fungal Surveillance Net (CHIF-NET) Study. J Clin Microbiol. 2018;56(7): DOI: 10.1128/JCM.00577-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Xiao M, Fan X, Chen SC, et al. Antifungal susceptibilities of Candida glabrata species complex, Candida krusei, Candida parapsilosis species complex and Candida tropicalis causing invasive candidiasis in China: 3 year national surveillance. J Antimicrob Chemother. 2015;70(3):802–810. DOI: 10.1093/jac/dku460 [DOI] [PubMed] [Google Scholar]
- [81].Wang D, An N, Yang Y, et al. Candida tropicalis distribution and drug resistance is correlated with ERG11 and UPC2 expression. Antimicrob Resist Infect Control. 2021;10(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Wang Y, Fan X, Wang H, et al. Continual decline in Azole susceptibility rates in Candida tropicalis over a 9-year period in China. Front Microbiol. 2021;12:702839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Jin L, Cao Z, Wang Q, et al. MDR1 overexpression combined with ERG11 mutations induce high-level fluconazole resistance in Candida tropicalis clinical isolates. BMC Infect Dis. 2018;18(1):162. DOI: 10.1186/s12879-018-3082-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Jiang C, Dong D, Yu B, et al. Mechanisms of azole resistance in 52 clinical isolates of Candida tropicalis in China. J Antimicrob Chemother. 2013;68(4):778–785. DOI: 10.1093/jac/dks481 [DOI] [PubMed] [Google Scholar]
- [85].Wang Q, Li C, Tang D, et al. Molecular epidemiology of Candida tropicalis isolated from urogenital tract infections. Microbiologyopen. 2020;9(11):e1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Fan X, Xiao M, Zhang D, et al. Molecular mechanisms of azole resistance in Candida tropicalis isolates causing invasive candidiasis in China. Clin Microbiol Infect. 2019;25(7):885–891. DOI: 10.1016/j.cmi.2018.11.007 [DOI] [PubMed] [Google Scholar]
- [87].Fan X, Xiao M, Wang H, et al. Multilocus sequence typing indicates diverse origins of invasive Candida tropicalis isolates in China. Chin Med J (Engl). 2014;127(24):4226–4234. [PubMed] [Google Scholar]
- [88].Wang Q, Tang D, Tang K, et al. Multilocus sequence typing reveals clonality of Fluconazole-Nonsusceptible Candida tropicalis: a Study from Wuhan to the Global. Front Microbiol. 2020;11:554249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Fan X, Xiao M, Liao K, et al. Notable increasing trend in Azole non-susceptible Candida tropicalis causing invasive Candidiasis in China (August 2009 to July 2014): molecular epidemiology and clinical Azole consumption. Front Microbiol. 2017;8:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Hou X, Xiao M, Wang H, et al. Profiling of PDR1 and MSH2 in Candida glabrata bloodstream isolates from a multicenter study in China. Antimicrob Agents Chemother. 2018;62(6): DOI: 10.1128/AAC.00153-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Yang W, Ji X. Analysis of the microbial species, antimicrobial sensitivity and drug resistance in 2652 patients of nursing hospital. Heliyon. 2020;6(5):e03965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Yao D, Chen J, Chen W, et al. Mechanisms of azole resistance in clinical isolates of Candida glabrata from two hospitals in China. Infect Drug Resist. 2019;12:771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Hou X, Xiao M, Chen SC, et al. Molecular epidemiology and antifungal susceptibility of Candida glabrata in China (August 2009 to July 2014): a multi-center study. Front Microbiol. 2017;8:880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Liu Y, Kang M, Ye H, et al. Analysis on clinical characteristics and drug resistance of Candida parapsilosis bloodstream infections in West China Hospital, China, from 2012 to 2015. Journal de Mycologie Medicale. 2018;28(1):222–226. [DOI] [PubMed] [Google Scholar]
- [95].Zhang L, Yu SY, Chen SC, et al. Molecular Characterization of Candida parapsilosis by microsatellite typing and emergence of clonal antifungal drug resistant strains in a multicenter surveillance in China. Front Microbiol. 2020;11:1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Feng W, Yang J, Wang Y, et al. ERG11 mutations and upregulation in clinical itraconazole-resistant isolates of Candida krusei. Can J Microbiol. 2016;62(11):938–943. [DOI] [PubMed] [Google Scholar]
- [97].Gong J, Xiao M, Wang H, et al. Genetic differentiation, diversity, and drug susceptibility of Candida krusei. Front Microbiol. 2018;9:2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Cheng JW, Yu SY, Xiao M, et al. Identification and antifungal susceptibility profile of Candida guilliermondii and Candida fermentati from a multicenter study in China. J Clin Microbiol. 2016;54(8):2187–2189. DOI: 10.1128/JCM.00938-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Cheng JW, Liao K, Kudinha T, et al. Molecular epidemiology and azole resistance mechanism study of Candida guilliermondii from a Chinese surveillance system. Sci Rep. 2017;7(1):907. DOI: 10.1038/s41598-017-01106-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Tian S, Bing J, Chu Y, et al. Genomic epidemiology of Candida auris in a general hospital in Shenyang, China: a three-year surveillance study. Emerging Microbes Infect. 2021;10(1):1088–1096. DOI: 10.1080/22221751.2021.1934557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Tian S, Rong C, Nian H, et al. First cases and risk factors of super yeast Candida auris infection or colonization from Shenyang, China. Emerging Microbes Infect. 2018;7(1):128. DOI: 10.1038/s41426-018-0131-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Hou X, Xiao M, Chen SC, et al. Identification and antifungal susceptibility profiles of Candida haemulonii species complex clinical isolates from a multicenter study in China. J Clin Microbiol. 2016;54(11):2676–2680. DOI: 10.1128/JCM.01492-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Mixão V, Gabaldón T. Genomic evidence for a hybrid origin of the yeast opportunistic pathogen Candida albicans. BMC Biol. 2020;18(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Sardi JCO, Scorzoni L, Bernardi T, et al. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol. 2013;62(Pt 1):10–24. [DOI] [PubMed] [Google Scholar]
- [105].Verma R, Pradhan D, Hasan Z, et al. A systematic review on distribution and antifungal resistance pattern of Candida species in the Indian population. Med Mycol. 2021;59(12):1145–1165. [DOI] [PubMed] [Google Scholar]
- [106].Zhang Z, Bai HH, Wang FJ, et al. Analysis of homology and drug sensitivity of vaginal isolates of 10 patients with recurrent vulvovaginal candidiasis in recurrent episodes. Zhonghua Fu Chan Ke Za Zhi. 2020;55(3):177–182. DOI: 10.3760/cma.j.cn112141-20191210-00665 [DOI] [PubMed] [Google Scholar]
- [107].Pappas PG, Lionakis MS, Arendrup MC, et al. Invasive candidiasis. Nat Rev Dis Primers. 2018;4:18026. [DOI] [PubMed] [Google Scholar]
- [108].Tsai MH, Hsu JF, Yang LY, et al. Candidemia due to uncommon Candida species in children: new threat and impacts on outcomes. Sci Rep. 2018;8(1):15239. DOI: 10.1038/s41598-018-33662-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Chow NA, Gade L, Tsay SV, et al. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis. 2018;18(12):1377–1384. DOI: 10.1016/S1473-3099(18)30597-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Afsarian MH, Badali H, Boekhout T, et al. Multilocus sequence typing of Candida albicans isolates from a burn intensive care unit in Iran. J Med Microbiol. 2015;64(Pt 3):248–253. [DOI] [PubMed] [Google Scholar]
- [111].Chapman B, Slavin M, Marriott D, et al. Changing epidemiology of candidaemia in Australia. J Antimicrob Chemother. 2017;72(4):1103–1108. DOI: 10.1093/jac/dkx047 [DOI] [PubMed] [Google Scholar]
- [112].Lamoth F, Lockhart SR, Berkow EL, et al. Changes in the epidemiological landscape of invasive candidiasis. J Antimicrob Chemother. 2018;73(suppl_1):i4–i13. [DOI] [PubMed] [Google Scholar]
- [113].Hachem R, Hanna H, Kontoyiannis D, et al. The changing epidemiology of invasive candidiasis: candida glabrata and Candida krusei as the leading causes of candidemia in hematologic malignancy. Cancer. 2008;112(11):2493–2499. [DOI] [PubMed] [Google Scholar]
- [114].Gupta AK, Venkataraman M. Antifungal resistance in Superficial mycoses. J Dermatolog Treat. 2021;32:1–25. [DOI] [PubMed] [Google Scholar]
- [115].Pristov KE, Ghannoum MA. Resistance of Candida to azoles and echinocandins worldwide. Clin Microbiol Infect. 2019;25(7):792–798. [DOI] [PubMed] [Google Scholar]
- [116].Whaley SG, Berkow EL, Rybak JM, et al. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front Microbiol. 2016;7:2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Corcione S, D’Avolio A, Pasero D, et al. Acquisition of FKS2 mutation after echinocandin treatment of infective endocarditis by Candida glabrata. Le Infezioni in Medicina. 2019;27(3):328–331. [PubMed] [Google Scholar]
- [118].Forsberg K, Woodworth K, Walters M, et al. Candida auris: the recent emergence of a multidrug-resistant fungal pathogen. Med Mycol. 2019;57(1):1–12. DOI: 10.1093/mmy/myy054 [DOI] [PubMed] [Google Scholar]
- [119].Alastruey-Izquierdo A, Melhem MS, Bonfietti LX, et al. Susceptibility test for fungi: clinical and laboratorial correlations in medical mycology. Rev Inst Med Trop Sao Paulo. 2015;57 Suppl 19(Suppl 19):57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Badiee P, Badali H, Boekhout T, et al. Antifungal susceptibility testing of Candida species isolated from the immunocompromised patients admitted to ten university hospitals in Iran: comparison of colonizing and infecting isolates. BMC Infect Dis. 2017;17(1):727. DOI: 10.1186/s12879-017-2825-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Briano F, Magnasco L, Sepulcri C, et al. Candida auris Candidemia in critically Ill, colonized patients: cumulative incidence and risk factors. Infect Dis Ther. 2022;11(3):1149–1160. DOI: 10.1007/s40121-022-00625-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Lau AF, Kabir M, Chen SC, et al. Candida colonization as a risk marker for invasive candidiasis in mixed medical-surgical intensive care units: development and evaluation of a simple, standard protocol. J Clin Microbiol. 2015;53(4):1324–1330. DOI: 10.1128/JCM.03239-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr. 2016;4(2). DOI: 10.1128/microbiolspec.VMBF-0016-2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Sjölund M, Bonnedahl J, Hernandez J, et al. Dissemination of multidrug-resistant bacteria into the Arctic. Emerg Infect Dis. 2008;14(1):70–72. DOI: 10.3201/eid1401.070704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Dynowska M, Ejdys E, Biedunkiewicz A, et al. Yeasts isolated from frequently in-patients and out-patients. Ann Parasitol. 2014;60(3):199–206. [PubMed] [Google Scholar]
- [126].Diallo OO, Baron SA, Abat C, et al. Antibiotic resistance surveillance systems: a review. J Glob Antimicrob Resist. 2020;23:430–438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting our study can be provided by demand from the corresponding author (email: zeng_yb@163.com) or the first author (email: bilal.microbiologist@yahoo.com).


