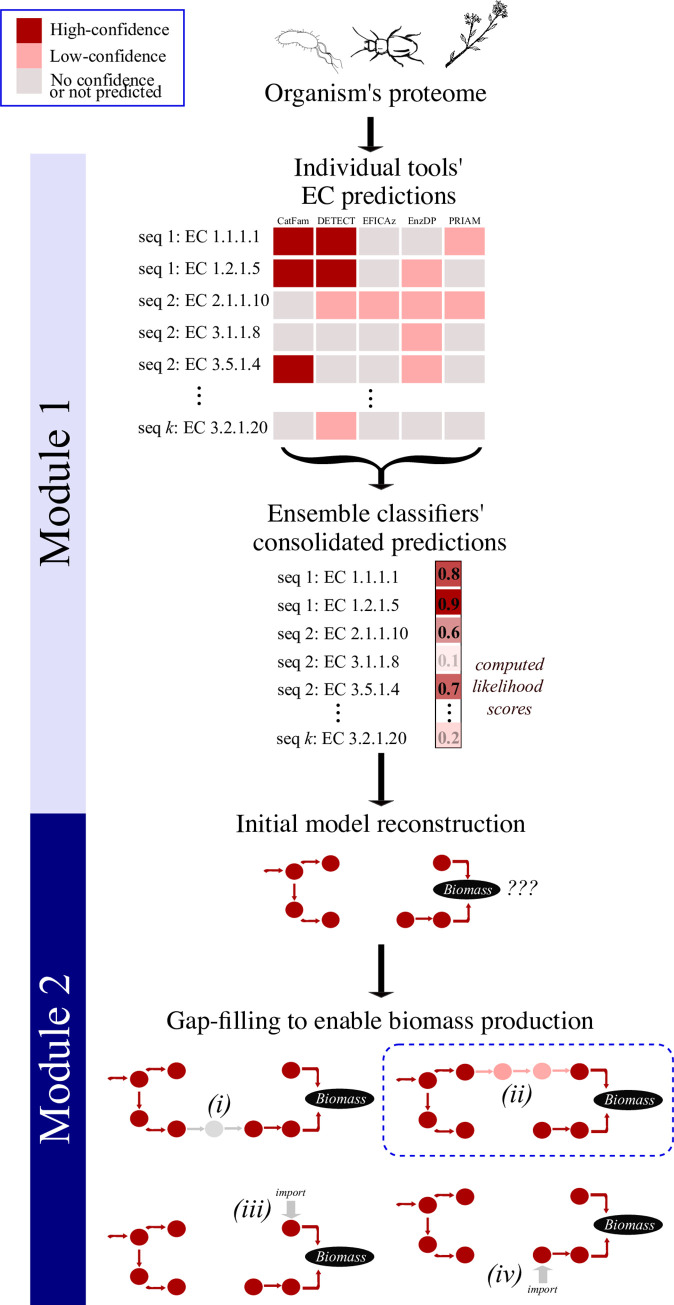
Fig 1. Overview of Architect’s methodology.
Given an organism’s protein sequences, Architect first runs 5 enzyme annotation tools, then computes a likelihood score for each annotation using an ensemble approach (module 1). From high-confidence EC predictions, Architect reconstructs a high-confidence metabolic model which it then gap-fills to enable biomass production using the aforementioned confidence scores (module 2). In the illustrated example, 4 sets of reactions are considered for gap-filling, with the solution highlighted in the blue box yielding the highest score.