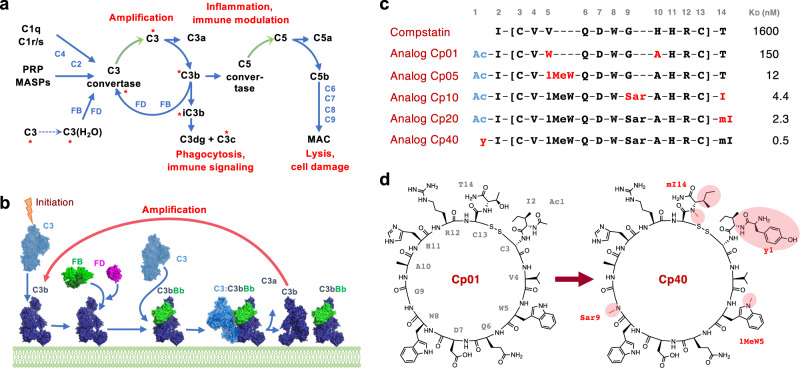
Fig. 1. Overview of complement mechanisms and compstatin-based inhibitor development.
a Simplified scheme of the complement cascade with major effector functions. Molecular targets of compstatin are marked with an asterisk. b Structural model of the amplification loop based on the crystal structures of C3 (2A73)22, C3b (2I07)18, FB (2OK5)50, FD (2XW9)5, C3b2Bb2SCIN2 (2WIN)6, and C3a (4HW5)51. c Amino acid sequences and target affinities of compstatin and major analogs relevant for this study. Residue numbers are indicated at the top, and changes from the previous analog are highlighted in red. Binding affinities (KD) to C3/C3b determined by surface plasmon resonance (SPR) are shown. d Comparison between the chemical structures of analogs Cp01 and Cp40, wherein modifications are highlighted in red. Abbreviations: Ac, Acetyl; FB, factor B; FD, factor D; MAC, membrane attack complex; MASPs, mannose binding lectin-associated serine proteases; PRP, pattern recognition protein. Reprinted from7 Trends in Pharmacological Sciences, Vol 8, C. Lamers, D.C. Mastellos, D. Ricklin, J.D. Lambris, Compstatins: the dawn of clinical C3-targeted complement inhibition, 629–640, Copyright 2022, with permission from Elsevier.