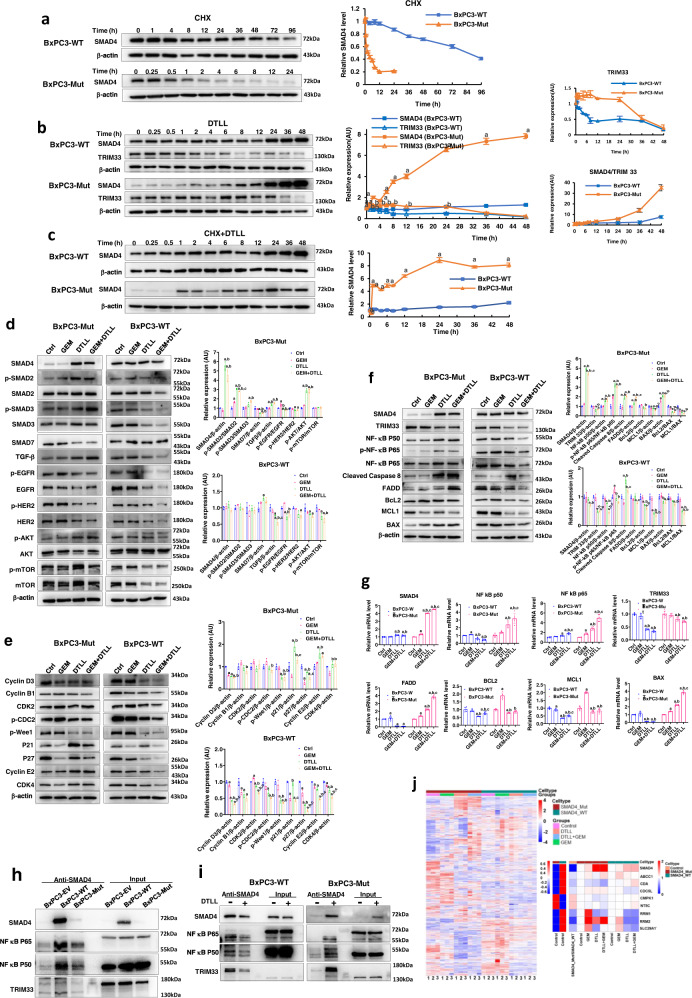
Fig. 8. Different mechanisms of the gemcitabine response in BxPC-3 cells affected by wild-type and mutant SMAD4 along with DTLL.
a The protein levels and half-life times of wild-type and mutant SMAD4 in BxPC3-WT and BxPC3-Mut stably transfected cells. Data are shown as the mean ± SD from three biologically independent samples (n = 3). b Different SMAD4 and TRIM33 levels of mutant and wild-type SMAD4 with DTLL induction in BxPC3-WT and BxPC3-Mut stably transfected cells. Data are shown as the mean ± SD from three biologically independent samples (n = 3). ‘a’ indicates p < 0.05 with comparison SMAD4 of BxPC3-Mut versus BxPC3-WT cells, ‘b’ represents p < 0.05 with comparison TRIM33 of AsPC-1 versus MIA PaCa-2 cells. When treated with DTLL, p < 0.001 for SMAD4 at all time points except for 0, 0.5, and 1 h, p < 0.001 for TRIM33 at 1, 2, 6, 8, 12, and 24 h. c Distinct protein levels and half-life times of mutant and wild-type SMAD4 with DTLL induction. Data are shown as the mean ± SD from three biologically independent samples (n = 3). ‘a’ indicates p < 0.05 with comparison of BxPC3-Mut versus BxPC3-WT cells. When treated with CHX + DTLL, p < 0.001 for SMAD4 at all time points except for 0, 0.25 and 0.5 h. d Different cell signaling pathways affected by mutant and wild-type SMAD4 with DTLL induction. Data are shown as the mean ± SD from three biologically independent samples (n = 3). In BxPC3-Mut cells, p < 0.001 (‘GEM + DTLL’versus Control) for SMAD4, p-SMAD2/SMAD2, SMAD7, TGFβ and p-AKT/AKT; p < 0.001 (‘GEM + DTLL’versus GEM) for SMAD4, p-SMAD2/SMAD2, TGFβ, p-EGFR/EGFR and p-AKT/AKT; p < 0.001 (DTLL versus Control) for SMAD4, p-SMAD2/SMAD2, SMAD7 and p-AKT/AKT; p < 0.001 (DTLL versus GEM) for SMAD4, p-SMAD2/SMAD2, p-EGFR/EGFR and p-AKT/AKT. In BxPC3-WT cells, p < 0.001 (‘GEM + DTLL’versus Control) for TGFβ; p < 0.001 (DTLL versus GEM) for p-EGFR/EGFR. e Cell cycle-related proteins were determined by Western blot assays in BxPC3-Mut and BxPC3-WT stably transfected cells after treatments with 2 µM gemcitabine, 0.1 nM DTLL or its combination at 37 °C for 4 h. Data are shown as the mean ± SD from three biologically independent samples (n = 3). In BxPC3-Mut cells, p < 0.001 (‘GEM + DTLL’versus Control) for Cyclin D3 and p27; p < 0.001 (DTLL versus Control) for Cyclin D3 and p21; p < 0.001 (DTLL versus GEM) for p21 and p27. In BxPC3-WT cells, p < 0.001 (‘GEM + DTLL’versus Control) for Cyclin D3, Cyclin B1, CDK2, p-CDC2, p-Weel, p21, p27, Cyclin E2 and CDK4; p < 0.001 (‘GEM + DTLL’versus GEM) for Cyclin D3, CDK2, p-Weel, p27, Cyclin E2 and CDK4; p < 0.001 (‘GEM + DTLL’versus DTLL) for CDK2, p21, Cyclin E2 and CDK4; p < 0.001 (DTLL versus Control) for Cyclin D3, CDK2, p-Weel, p27, Cyclin E2 and CDK4; p < 0.001 (DTLL versus GEM) for Cyclin D3, CDK2, p21and p27; p < 0.001 (GEM versus Control) for CDK2, p21, p27and Cyclin E2. f TRIM33, NF-κB and apoptosis-related proteins were determined by Western blot assays in BxPC3-Mut and BxPC3-WT stably transfected cells after treatment with gemcitabine and DTLL. Data are shown as the mean ± SD from three biologically independent samples (n = 3). In BxPC3-WT cells, p < 0.001 (‘GEM + DTLL’versus Control) for SMAD4, NF-kB p50, p-NF-kB p65/NF-kB p65, Cleaved Caspase 8, FADD, MCL1, BAX, BCL2/BAX and MCL1/BAX; p < 0.001 (‘GEM + DTLL’versus GEM) for SMAD4, NF-kB p50, p-NF-kB p65/NF-kB p65, Cleaved Caspase 8, FADD; p < 0.001 (‘GEM + DTLL’versus DTLL) for SMAD4 and FADD; p < 0.001 (DTLL versus Control) for SMAD4, p-NF-kB p65/NF-kB p65, Cleaved Caspase 8, BAX, BCL2/BAX and MCL1/BAX; p < 0.001 (DTLL versus GEM) for SMAD4, p-NF-kB p65/NF-kB p65 and Cleaved Caspase 8; p < 0.001 (GEM versus Control) for p-NF-kB p65/NF-kB p65 and MCL1/BAX. In BxPC3-WT cells, p < 0.001 (‘GEM + DTLL’versus Control) for TRIM33, NF-kB p50, BCL2, MCL1, and MCL1/BAX; p < 0.001 (‘GEM + DTLL’versus GEM) for NF-kB p50, BCL2, MCL1 and MCL1/BAX; p < 0.001 (‘GEM + DTLL’versus DTLL) for NF-kB p50; p < 0.001 (DTLL versus Control) for Cleaved Caspase 8, MCL1, and MCL1/BAX; p < 0.001 (DTLL versus GEM) for Cleaved Caspase 8 and MCL1. g The mRNA levels of SMAD4, TRIM33, P50, P65, FADD, Bcl-2, MCL1, and BAX were determined in BxPC3-Mut and BxPC3-WT cells after treatments with gemcitabine, DTLL and their combination by using qRT-PCR. Data are shown as the mean ± SD from three biologically independent samples (n = 3). In BxPC3-WT cells, p < 0.001 (‘GEM + DTLL’versus Control) for NF-kB p50, NF-kB p65, TRIM 33, FADD, BcL2, MCL1, and Bax; p < 0.001 (‘GEM + DTLL’versus GEM) for NF-kB p50, NF-kB p65, TRIM 33, MCL1and Bax; p < 0.001 (‘GEM + DTLL’versus DTLL) for NF-kB p65 and MCL1; p < 0.001 (DTLL versus Control) for SMAD4, NF-kB p50, NF-kB p65, TRIM 33, BcL2, MCL1, and Bax; p < 0.001 (DTLL versus GEM) for SMAD4, NF-kB p50, TRIM 33, BcL2, MCL1, and Bax. In BxPC3-Mut cells, p < 0.001 (‘GEM + DTLL’versus Control) for SMAD4, NF-kB p50, NF-kB p65, TRIM 33, FADD and Bax; p < 0.001 (‘GEM + DTLL’versus GEM) for SMAD4, NF-kB p50, NF-kB p65, TRIM 33, FADD, BcL2, MCL1, and Bax; p < 0.001 (‘GEM + DTLL’versus DTLL) for SMAD4, NF-kB p65, FADD, and Bax; p < 0.001 (DTLL versus Control) for SMAD4, NF-kB p50, NF-kB p65, FADD, and Bax; p < 0.001 (DTLL versus GEM) for SMAD4, NF-kB p50, NF-kB p65, FADD, BcL2, MCL1, and Bax; p < 0.001 (GEM versus Control) for SMAD4, FADD, BcL2, MCL1, and Bax. h Specific interactions of SMAD4 with NF-κB and TRIM33 in BxPC3-EV, BxPC3-WT, and BxPC3-Mut stably transfected cells. The results were obtained from three biologically independent experiments. i Different protein interactions of mutant or wild-type SMAD4 with DTLL induction in BxPC3-WT and BxPC3-Mut stably transfected cells. The results were obtained from three biologically independent experiments. j Different proteomic and gemcitabine pharmacokinetic profiles in mutant and wild-type SMAD4 cells after treatment with GEM, DTLL or both. Data are shown as the mean ± SD from three biologically independent samples (n = 3). For d–g, ‘a’ indicates p < 0.05 as compared with the control, ‘b’, p < 0.05 compared with the GEM group and ‘c’, p < 0.05 compared with the DTLL group. One-way ANOVA with Bonferroni post hoc test was used in d–g; Paired-samples t-test was two-sided and used in b and c. All specific p values are presented in the Source Data file and only significant values are shown in figures. Source data are provided in the Source Data file.