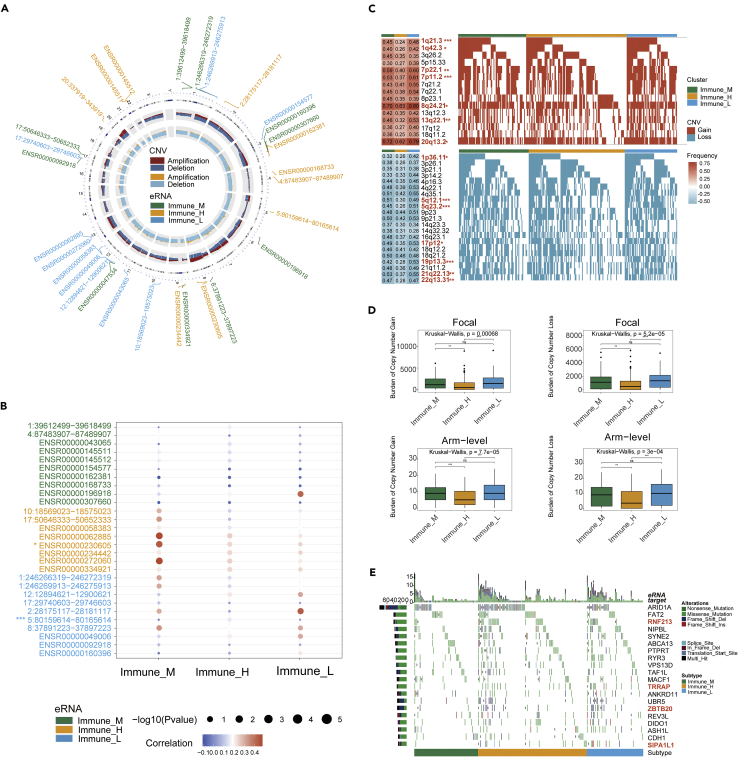
Figure 4.
Genomic mutation changes affect eRNA expression
(A) The circle plot showed a distribution of 30 eRNAs (top ten differentially expressed in three eRNA subtypes, respectively). The circles from outside to inside stand for chromosome location, copy-number frequency (dark red: amplification, dark blue: deletion), and copy number gistic score (yellow: amplification, blue: deletion), respectively. Each green font showed an Immune_M differentially expressed eRNA, each yellow font showed an Immune_H differentially expressed eRNA, and each blue font showed an Immune_L differentially expressed eRNA.
(B) The bubble plot displayed the Spearman’s correlation between the copy number alterations and the expression of differential eRNAs. Asterisks (∗) in the upper left corner of eRNAs indicate significant differences in CNV distributions between three eRNA subtypes. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(C) Detailed plots with copy number gain (up) and copy number loss (down) between three eRNA subtypes. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) Distribution of focal and broad copy number alterations between three eRNA subtypes. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns = not statistically significant.
(E) Mutational landscape of target gene between three eRNA subtypes.