Figure 3.
Computational design and structure validation of 9–12 amino acid macrocycles
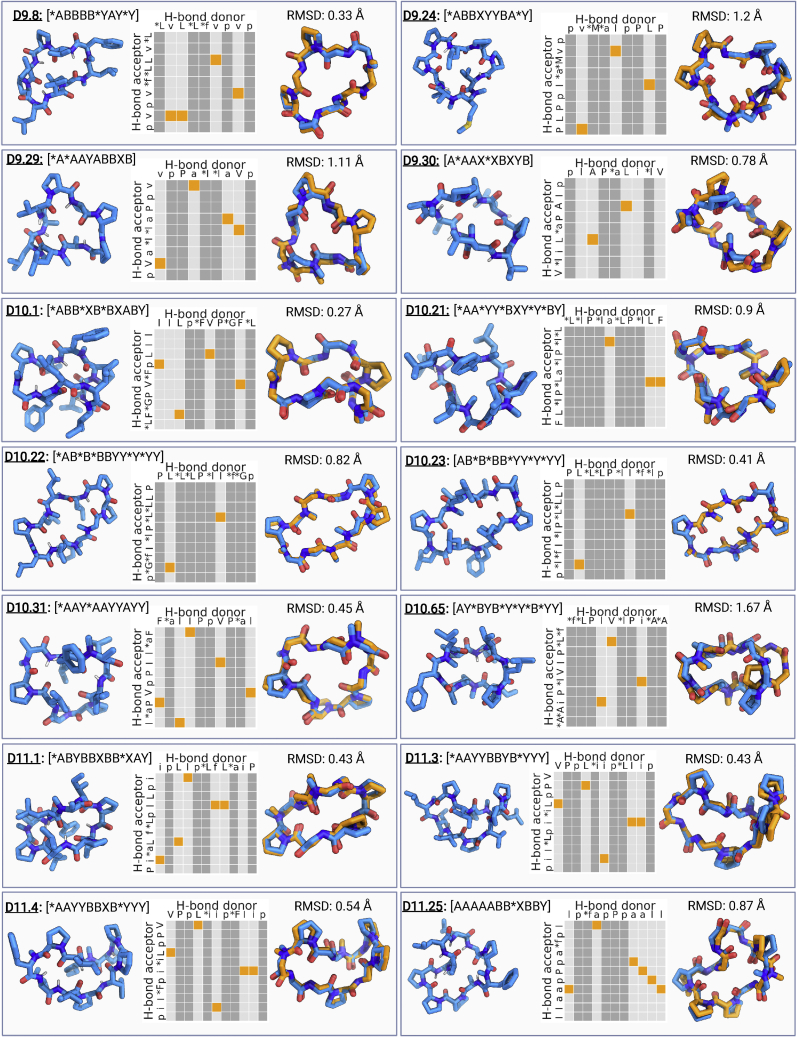
Structural validation of computationally designed macrocycles. Each panel shows the design model and torsion bin string describing the design model (left), hydrogen bonding pattern for the design model (middle), and superposition between the design model (blue) and the X-ray structure (orange) (right). For hydrogen bonding graphs, the orange boxes highlight the designed intramolecular hydrogen bonds. Amino acids without a backbone hydrogen bond donor (proline, D-proline, and N-methylated amino acids) are marked by darker gray columns. Side chains for non proline residues not shown for clarity in the superposition graphs. RMSD between the design model and X-ray structure was calculated over all the backbone heavy atoms (C, CA, N, O, and CN). ∗ in macrocycle sequence denotes the N-methylated amino acid positions and lowercase denotes the D-amino acids.
See also Table S2, Figures S1, S3, and S5, and Data S1 and S2.