Figure S1.
Design and selection of membrane-permeable peptides, related to Figures 1 and 3 and STAR Methods
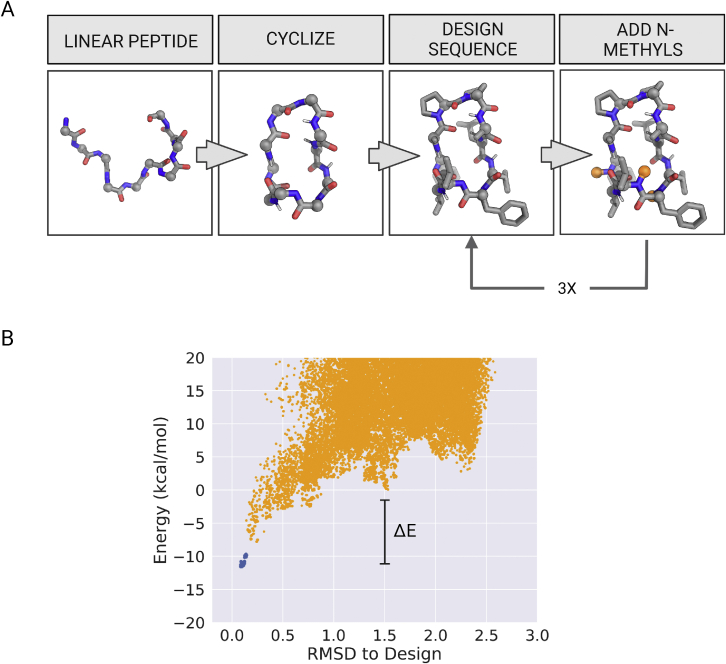
(A) Overall schematic of the in silico pipeline for the design of membrane-permeable peptides. Design process starts with a linear polyglycine peptide chain that is cyclized using Rosetta generalized kinematic closure (genKIC) protocol. Iterative rounds of amino acid sequence design and N-methylation of non-hydrogen-bonded NH groups are performed to design low-energy macrocycles with no unsatisfied backbone NH groups. The process is repeated to sample 105–106 design models that are clustered to identify permeable macrocycles with diverse shapes and sizes.
(B) An example energy versus RMSD to design plot from structure prediction runs using Rosetta simple_cycpep_predict application. Diverse conformations for a given amino acid sequence are generated using generalized kinematic closure (genKIC) protocol and energy-minimized using Rosetta FastRelax protocol. Each orange point represents an independently predicted structure. Blue dots represent the local minimization of the designed macrocycle structure. Landscapes that funnel into the design structure as the lowest energy structure and have a big energy gap (ΔE) between the designed fold and other unfolded states are selected for experimental characterization.