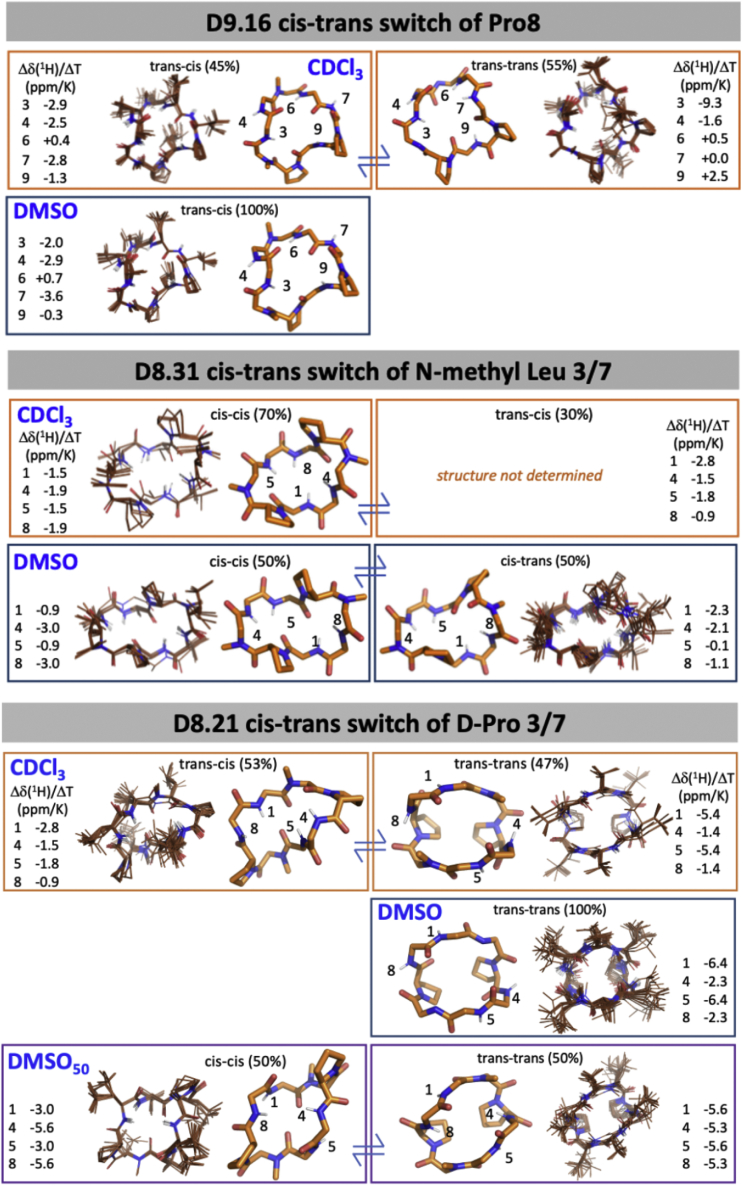
Figure S6.
NMR structures in different solvents, related to Figure 4
NMR-derived structures in the indicated solvents d6-DMSO, CDCl3, or 50:50 d6-DMSO/H2O (DMSO50) are shown for D9.16, D8.31, and D8.21. The overlay structure of the ensemble of 20 lowest energy structures is shown along with the backbone structure of the medoid conformation with NH protons labeled. The amide proton temperature coefficients Δδ(1H)/ΔT are given for each of the HN resonances in each conformation. Less negative coefficients indicate increased hydrogen bond propensity and correlate with hydrogen bonds in the structures. In general, upon increasing temperature, amide 1H chemical shifts move upfield, which is attributed to a lengthening of the hydrogen bond and decreased shielding from the hydrogen bond acceptor (Baxter and Williamson, 1997). Large changes in chemical shift give large negative temperature coefficients and indicate solvent exposed or weakly hydrogen-bonded NHs. It was empirically found that for proteins in aqueous solution that amide protons with Δδ(1H)/ΔT that are more positive than −4.6 ppm/K (less negative and even sometimes positive) are indicative of intramolecular hydrogen bonds (Cierpicki and Otlewski, 2001). For cyclic peptides, temperature coefficients have been used as a measure of hydrogen bonding potential and correlated with MD simulations and predicted structures in aqueous solution as well as in chloroform and DMSO (Wang et al., 2015). In most cases, the NOESY or ROESY data with the 3Jhnha coupling-derived dihedral restraints was sufficient to give a converged structure for the peptides. However, in the case of the trans-trans variant of D8.21, the symmetry and the more open conformation with fewer NOEs did not give a unique conformational solution. In these cases, the conformation that best correlated the temperature coefficients was selected.