Figure 1.
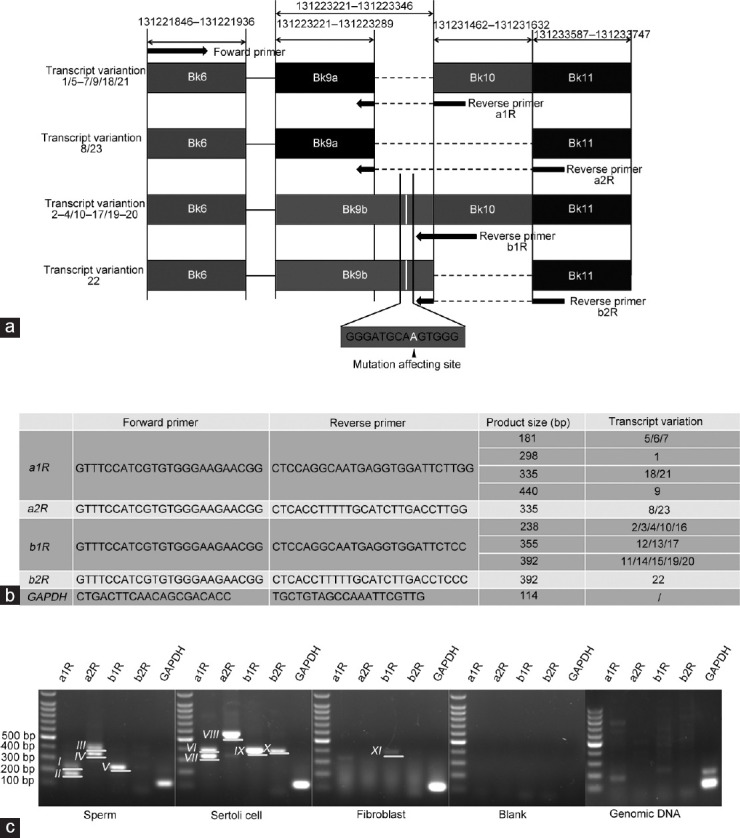
PCR detection of the transcripts that could be affected by the mutation in sperm. (a) The positions of the primers designed to distinguish the ODF2 transcripts with or without the sequence that could be affected by the mutation are indicated by the arrows. The mutated site is located in Bk9b (the location of the mutation affecting site is indicated in white, and sequence flanked the mutation affecting site is indicated below). The forward primer was shared. Reverse primer a1R and a2R were employed to detect transcripts containing Bk9a which does not contain the mutation affecting site. Reverse primer b1R and b2R were employed to detect transcripts containing Bk9b which contains the mutation affecting site. Only 4–5 nucleotides at the 3' terminal of these reverse primers match the sequence located in Bk9, indicating the annealing temperature is extremely low for these primers to bind to Bk9 directly unless incorporated with the rest of the primer sequences that match the sequence located in Bk10 or Bk11, avoiding the nonspecific amplification when common PCR conditions were used. (b) The primer sequences employed for detecting the transcripts. The product sizes of corresponding transcript variations are also listed. (c) PCR results for detecting the transcripts that could be affected by the mutation. cDNA derived from purified sperm was used as a template. cDNAs derived from Sertoli cells and fibroblast served as somatic controls. Water served as a negative control. Genomic DNA was also used as a PCR template for comparison, determining whether genomic DNA contamination existed in these cDNA specimens. The reverse primers used for each reaction are indicated above the lanes. GAPDH was detected as an internal control. It was confirmed that some of the transcripts that could be affected by the mutation were highly expressed in sperm (these transcripts generated product approximately 238 bp in size, numbered Band V), and the others were expressed in Sertoli cells and fibroblast (these transcripts generated products approximately 355 bp or 392 bp in size, numbered Band IX and Band X, respectively). Besides, unexpected products were generated (Band I, Band III, Band VI, and Band VIII. The sizes of these products do not match the expected ones), PCR amplification using genomic DNA as a template could not generate products with the same sizes as those generated from cDNA specimens, indicating there was no contamination of genomic DNA in these cDNA specimens. Bk: block, used for informal numbering of the exons in this work; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; cDNA: complementary DNA; ODF2: outer dense fiber protein 2.
