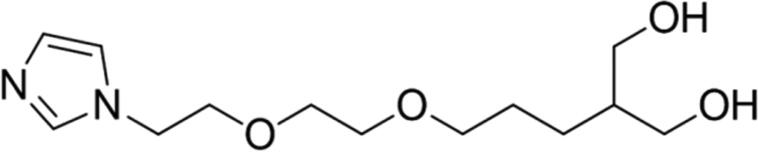
Table 1.
Molecular structures of ligands predicted through protonation analysis under physiologic pH (7.4).
| Prototype | Structurea | % of micro speciesa | Meet the druggability criteria of the Lipinski rule?b |
|---|---|---|---|
| 1 |
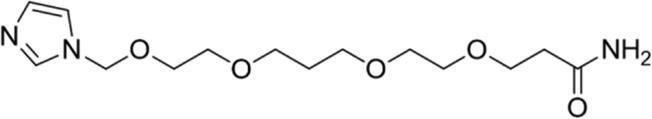
|
87.09 | Yes |
| 2 |
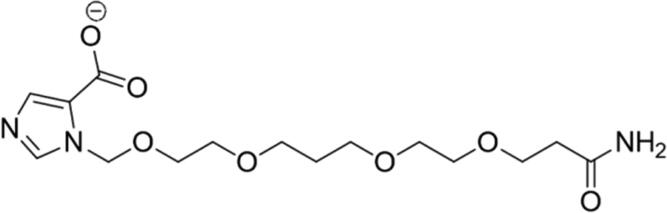
|
78.37 | Yes |
| 3 |
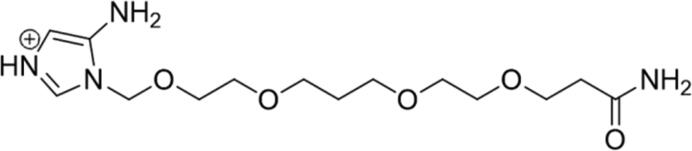
|
78.25 | Yes |
| 4 |
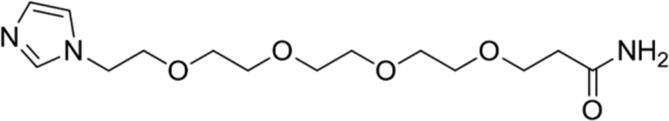
|
89.04 | Yes |
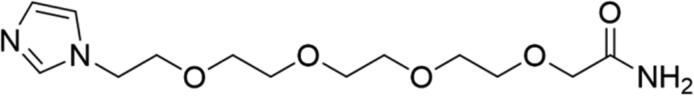
| 5 |
|
89.04 | Yes |
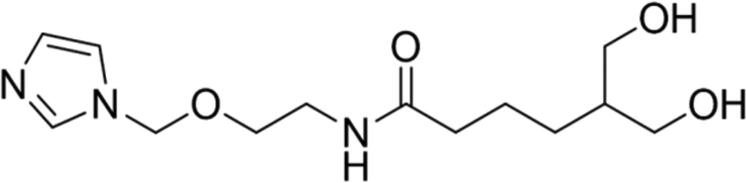
| 6 |
|
87.08 | Yes |
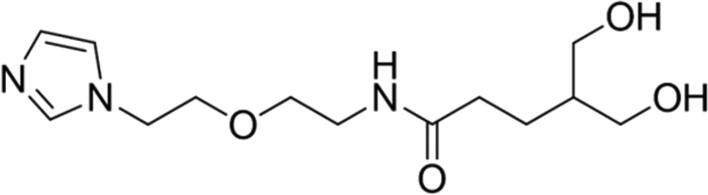
| 7 |
|
89.04 | Yes |
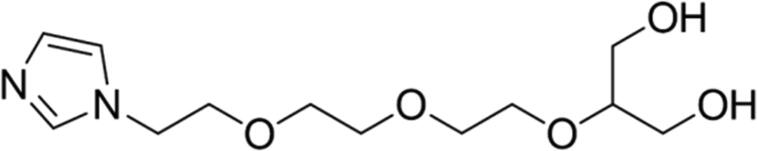
| 8 |
|
89.04 | Yes |
| 9 |
|
89.04 | Yes |
Calculated through Chemicalize web-based resource (https://chemicalize.com/welcome) (Swain, 2012) as the highest percentage of this micro species.
(Lipinski, 2004).
