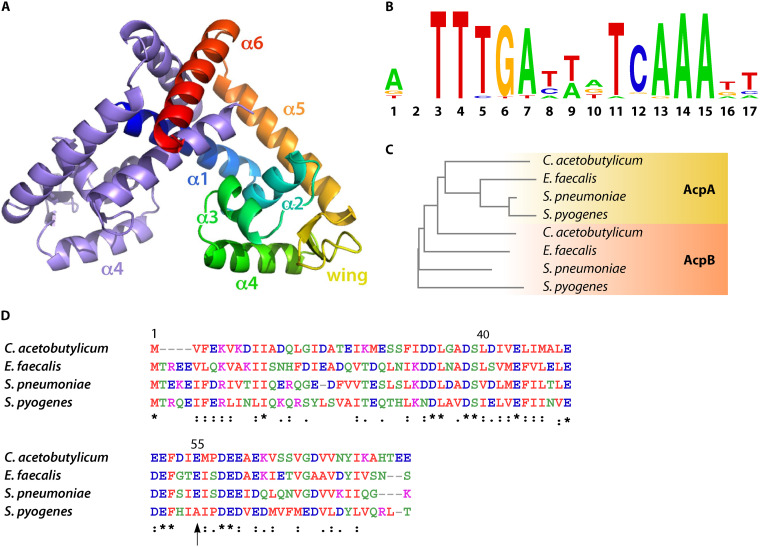
FIG 2.
Structural features of FabT, the ACP corepressor, and the FabT-DNA binding motif. (A) Overall structure of MarR family member dimer; one monomer is represented multicolored and the other in blue. Reprinted from reference 30 with permission. (B) FabT DNA binding motif; top, calculated from 56 FabT DNA binding sequences (https://regprecise.lbl.gov/regulog.jsp?regulog_id=3571). The relative size of letters corresponds to the relative frequency at which each nucleotide is present. (C) Phylogenetic neighbor-joining tree obtained from a multiple sequence alignment carried out using Clustal W (1.83). Branch lengths correspond to: C. acetobutylicum AcpA, 0.28257; E. faecalis AcpA, 0.19187; S. pneumoniae, AcpA 0.0325; S. pyogenes, AcpA 0.04858; C. acetobutylicum AcpB, 0.25594; E. faecalis AcpB, 0.29157; S. pneumonia, AcpB, 0.25143; S. pyogenes, AcpA, 0.35383. (D) AcpB alignment; multiple sequence alignment was carried out using Clustal W (1.83) (unpublished results), color code follows functional classification; *, identical in all four species: the same structural class; vertical arrow highlights amino acid residue 55, which is adjacent to the FabT-AcpB binding (32) but is not conserved in S. pyogenes.