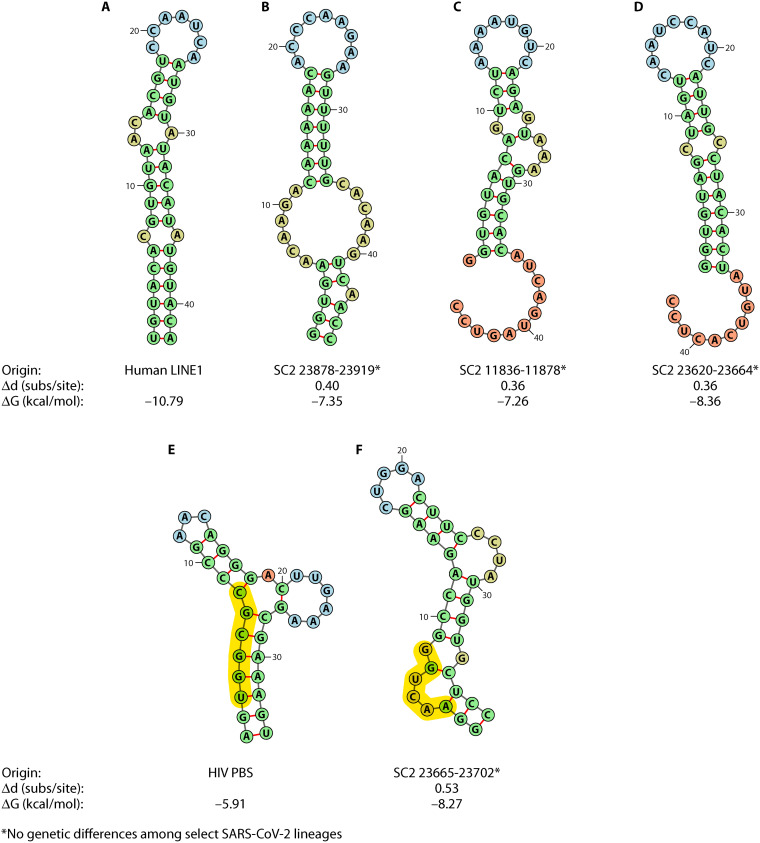
FIG 8.
Predicted RNA secondary structures for regions within the SARS-CoV-2 (SC2) genome sharing similar sequence content with original human LINE1 hairpin (200) (A) and HIV-1 tRNA annealing arm containing the primer-binding site (PBS) (B). Genomic coordinates within the SARS-CoV-2 reference genomes, including the original Wuhan-Hu-1 reference sequence (MN908947.3), USA-WA12020 (MN985325.1) as used in Zhang et al., and variants Alpha (MZ344997.1) and Delta (MZ359841.1), are provided (B, C, D, and F), as well as the Hamming distance for the alignment of the original sequence (A and E) with the reference SARS-CoV-2 sequence (Δd) and the minimum free energy value for predicted RNA secondary structure (ΔG) in RNAfold (138, 139). PBS sequence is highlighted in yellow. No genetic differences were observed between MZ344997.1, MZ359841.1, MN985325.1, and NC045512.2. A double GC base pair was added to each SARS-CoV-2 sequence (flanking) to force stem formation and is included in the pictured structure.