Figure 1.
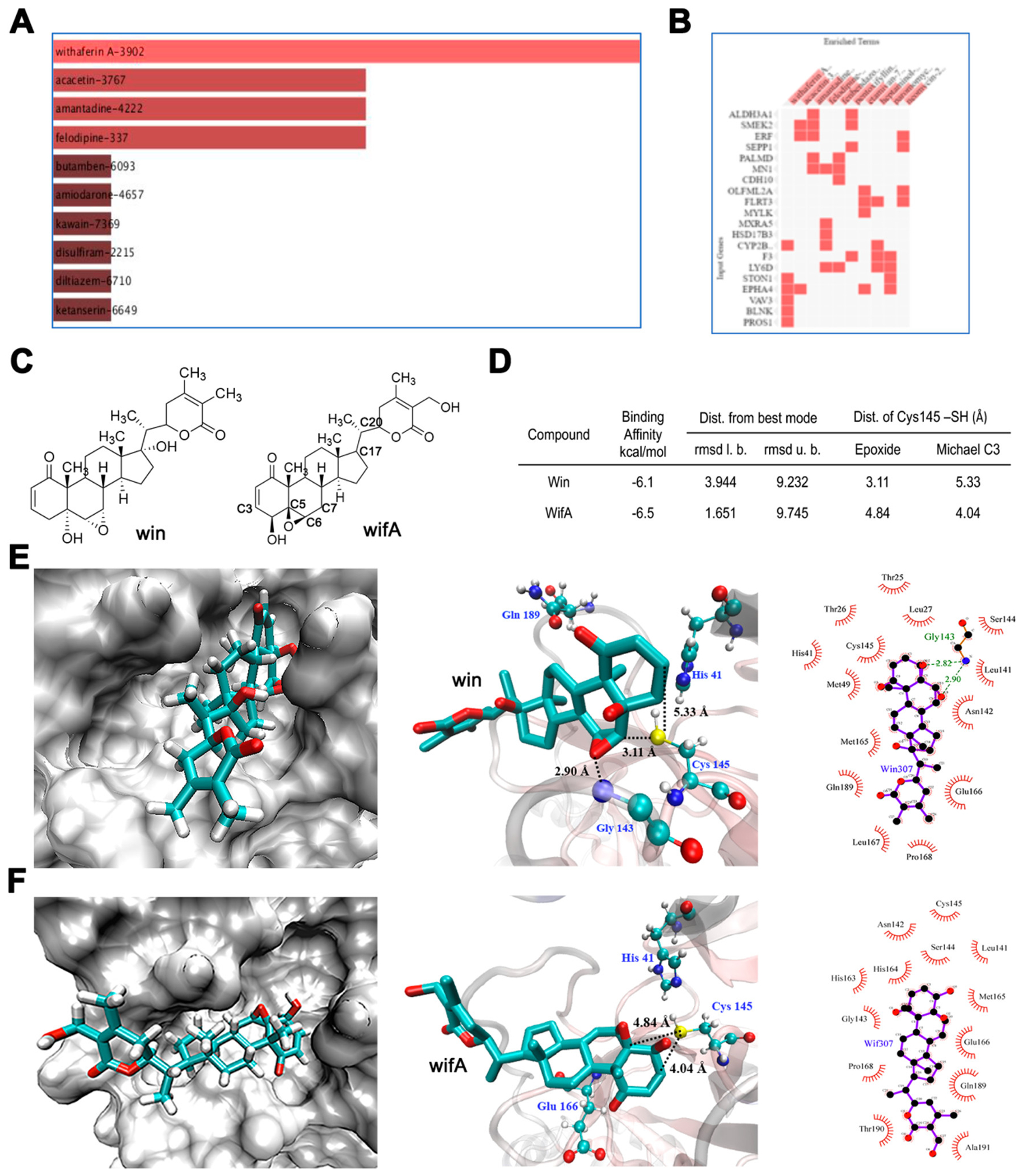
Identification of wifA and win as potential drug candidate against SARS-CoV-2. (A) Top drug candidates revealed by CMap analysis with genes uniquely upregulated in SARS-CoV-2-infected lung cells. (B) Genes associated with each drugs. In A and B all candidates shown are statistically significant at p < 0.05. (C) Structures of wifA and win. (D) Noncovalent binding of wifA and win at the active site of the SARS-CoV-2 cysteine protease Mpro using in silico docking studies. Binding parameters of wifA and win at the active site of Mpro. (E) Representative binding modes of win at the active site of Mpro in three different representations showing key distances and the interactions in Mpro-win. (F) Representative binding modes of wifA at the active site of Mpro in three different representations showing key distances and interactions in Mpro-wifA.
