Figure 4.
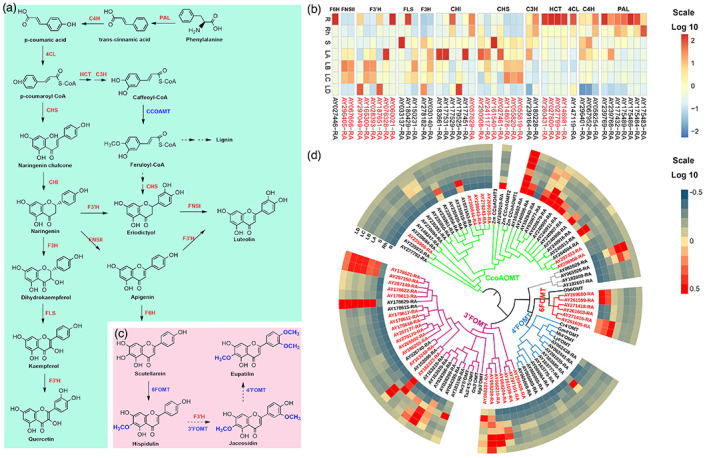
The identification and expression profiles of genes related to the biosynthesis of flavonoids in A. argyi. (a). The proposed flavonoid biosynthesis pathway in A. argyi. Red fonts represented the abbreviations of enzymes participating in the catalytic steps. And the full names of relative enzymes were shown in Data S9. (b). The expression patterns of candidate genes involved in flavonoids biosynthesis pathway in different tissues. The expanded genes were marked in red. R, root; Rh, rhizome; S, stem; LA, leaf buds, 0 day; LB, young leaves 15 days; LC, mature leaves 30 days; LD, old leaves 45 days. (c). Proposed biosynthesis pathways for hispidulin, jaceosidin and eupatilin. (d). Expression profile and phylogenetic tree of all members of the flavonoid O‐methyltransferase (FOMT) gene family in A. argyi. The genes in red were expanded genes. [Colour figure can be viewed at wileyonlinelibrary.com]
