Summary
The polyamine putrescine (1,4‐diaminobutane) contributes to cellular fitness in most organisms, where it is derived from the amino acids ornithine or arginine. In the chemical industry, putrescine serves as a versatile building block for polyamide synthesis. The green microalga Chlamydomonas reinhardtii accumulates relatively high putrescine amounts, which, together with recent advances in genetic engineering, enables the generation of a powerful green cell factory to promote sustainable biotechnology for base chemical production. Here, we report a systematic investigation of the native putrescine metabolism in C. reinhardtii, leading to the first CO2‐based bio‐production of putrescine, by employing modern synthetic biology and metabolic engineering strategies. A CRISPR/Cas9‐based knockout of key enzymes of the polyamine biosynthesis pathway identified ornithine decarboxylase 1 (ODC1) as a gatekeeper for putrescine accumulation and demonstrated that the arginine decarboxylase (ADC) route is likely inactive and that amine oxidase 2 (AMX2) is mainly responsible for putrescine degradation in C. reinhardtii. A 4.5‐fold increase in cellular putrescine levels was achieved by engineered overexpression of potent candidate ornithine decarboxylases (ODCs). We identified unexpected substrate promiscuity in two bacterial ODCs, which exhibited co‐production of cadaverine and 4‐aminobutanol. Final pathway engineering included overexpression of recombinant arginases for improved substrate availability as well as functional knockout of putrescine degradation, which resulted in a 10‐fold increase in cellular putrescine titres and yielded 200 mg/L in phototrophic high cell density cultivations after 10 days.
Keywords: amine oxidase, Chlamydomonas reinhardtii, CRISPR/Cas9, microalga, ornithine decarboxylase, polyamines
Introduction
Chemical synthesis of most commodity plastics is based on fossil petroleum feedstock, which could be readily replaced by bio‐produced monomers (Fiorentino et al., 2019). For example, synthesis of the polyamide Nylon‐4,6 requires the bio‐available monomers putrescine and adipic acid, offers excellent material properties and can be used in heavy‐duty fibre and engineering products (Roerdink and Warnier, 1985; Schneider and Wendisch, 2011). With up to 0.3% of the total biomass dry weight (BDW), putrescine is the most abundant endogenous polyamine (91 mol%) in C. reinhardtii (Lin and Lin, 2019), which makes it an ideal candidate for the development of a microbial green cell factory for polyamine production. Recently, we demonstrated great flexibility in the amino acid metabolism of this microalga by establishing engineered overproduction of the non‐native diamine cadaverine (Freudenberg et al., 2021).
The putrescine metabolism and its biological roles in Chlamydomonas reinhardtii are complex and require further investigations in order to develop advanced overproduction strategies. Putrescine possesses some nonspecific functions (derived from its chemical properties, e.g. positive charge at physiological pH), which include interactions with nucleic acids, phospholipids and proteins, thereby regulating transcription and translation (Igarashi and Kashiwagi, 2010; Michael, 2016a; Miller‐Fleming et al., 2015). In C. reinhardtii, it is assumed to be involved in nitrogen storage, cell cycle progression (Theiss et al., 2002), DNA secondary structure (G‐quadruplex) formation (Vinyard et al., 2018) and higher polyamine synthesis. In plants, polyamines have been shown to contribute to growth, development, cell signalling, photosynthesis as well as abiotic and biotic stress responses (Chen et al., 2019; Jancewicz et al., 2016; Poidevin et al., 2019; Rangan et al., 2014; Shu et al., 2012).
Similar to plants, putrescine biosynthesis in C. reinhardtii (Figure 1a) is intimately tied to nitrogen metabolism and arginine biosynthesis in the chloroplast (Harris et al., 2009). Whilst ammonium can be assimilated directly, environmental nitrate is initially reduced via nitrite to ammonium. The incorporation of inorganic ammonium into glutamate is catalysed by the glutamine synthetase/glutamate synthase cycle, which additionally requires α‐ketoglutarate from the tricarboxylic acid (TCA) cycle (Fernandez and Galvan, 2008). Nitrogen assimilation is amongst others regulated via feedback inhibition by excess arginine or ammonium, mediated by the NO‐inducible guanylate cyclase CYG56 (González‐Ballester et al., 2018). Putrescine synthesis starts with enzymatic acetylation and phosphorylation of glutamate, resulting in N‐acetylglutamate‐5‐phosphate. The enzyme for the second, rate‐limiting reaction (NAGK, N‐acetylglutamate kinase) is regulated via allosteric feedback inhibition by arginine (Slocum, 2005) and stimulation by the C/N state‐sensing PII signal transduction protein (Selim et al., 2020a; Zalutskaya et al., 2018). Subsequent ornithine biosynthesis from N‐acetylglutamate‐5‐phosphate requires another three enzymatic reactions, after which two possible pathways exist for putrescine synthesis, the ornithine and arginine decarboxylase (ODC and ADC) routes.
Figure 1.
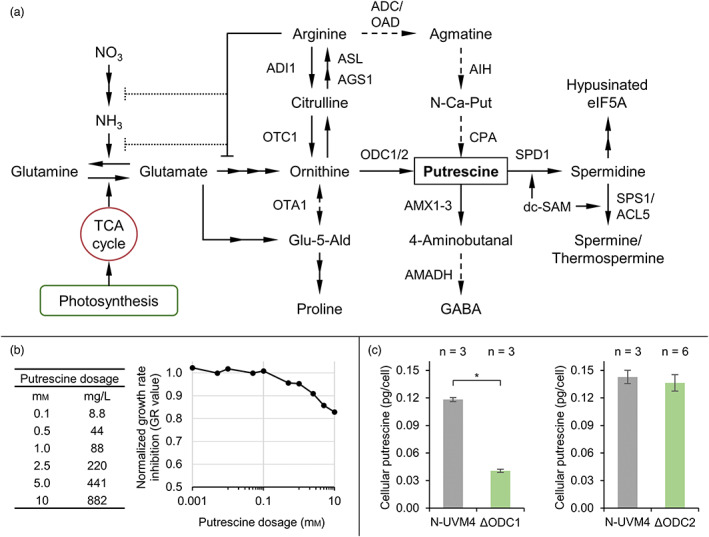
Investigating putrescine metabolism in Chlamydomonas reinhardtii. (a) Schematic of the putrescine metabolic network originating from the incorporation of inorganic carbon and nitrogen, leading to glutamate formation. Dashed lines indicate vestigial or hypothetic reactions. Multiple consecutive reactions are summarized by two or more arrows. An orthogonal bar at the end of a connection symbolizes direct (solid line) or indirect (dotted lines) feedback inhibition. (b) Effect of extracellular putrescine on cell growth, summarized as growth rate inhibition (GR value) of spiked UVM4 cultures normalized to the untreated control after 3 days. (c) Cellular putrescine levels during the early stationary growth phase in the N‐UVM4 strain compared to those in ΔODC1 and ΔODC2 knockout mutants. Error bars represent the standard deviation of biological replicates. Asterisks (*) indicate the significance level of an unpaired, two‐sided Student's t‐test assuming non‐homogenous variances (*P < 0.05). TCA cycle, Tricarboxylic acid cycle; ADI, Arginine deiminase; OTC, Ornithine carbamoyltransferase; ASL, Argininosuccinate lyase; AGS, Argininosuccinate synthase; ADC, Arginine decarboxylase; OAD, Arginine/ornithine decarboxylase; OTA, Ornithine aminotransferase; ODC, Ornithine decarboxylase; AIH, Agmatine iminohydrolase; CPA, N‐carbamoylputrescine amidohydrolase; AMX, Amine oxidase; AMADH, Aminoaldehyde dehydrogenase; SPD, Spermidine synthase; dc‐SAM, Decarboxylated S‐adenosyl methionine; eIF5A, Eukaryotic translation (initiation) factor 5A; SPS/ACL, (Thermo)spermine synthase; Glu‐5‐Ald, Glutamate‐5‐semialdehyde; GABA, γ‐aminobutyric acid; N‐Ca‐Put, N‐carbamoylputrescine. [Colour figure can be viewed at wileyonlinelibrary.com]
Chlamydomonas reinhardtii harbours an active ODC route (Figure 1a), which relies on the catalytic activity of ornithine decarboxylases, encoded by ODC1 and ODC2 (Merchant et al., 2007; Tassoni et al., 2018; Voigt et al., 2000). ODC1 expression is partially regulated by diurnal cycle‐dependent alternative splicing, whereby the functional mRNA variant (ODC1‐S) accumulates in the light phase (Labadorf et al., 2010; Pandey et al., 2020). Evidence suggests that ornithine is also substrate for ornithine aminotransferase, which catalyses the reaction towards glutamate‐5‐semialdehyde, thereby connecting putrescine and proline biosynthesis (Atteia et al., 2009; Zalutskaya et al., 2020).
The ADC route is present in most plants and few microalgae such as Chlorella vulgaris (Cohen et al., 1983; Michael, 2016b), but its activity in C. reinhardtii is unclear because the evidence for an arginine decarboxylase is lacking (Lin and Lin, 2019). This route requires arginine, which is synthesized by subsequent amidation of ornithine via carbamoyl phosphate and aspartate (Figure 1a). Arginine is decarboxylated by the activity of ADC to agmatine, which is converted to putrescine via N‐carbamoylputrescine.
Putrescine acts as an essential substrate for the synthesis of higher polyamines like spermidine, spermine or thermospermine. In the initial step, spermidine synthase (SPD1) catalyses the formation of spermidine, which is required for cell proliferation (Freudenberg et al., 2022). Putrescine is degraded by amine oxidases (AMX) to 4‐aminobutanal (Harris et al., 2009; Park et al., 2015), which could be oxidized to γ‐aminobutyric acid (GABA) by the activity of an aminoaldehyde dehydrogenase, as observed in Arabidopsis thaliana (Stiti et al., 2011). GABA might then be recycled into the TCA cycle via succinate. Other catabolic reactions such as N‐acetylation or N‐methylation of putrescine, which have been described in other organisms (Li et al., 2016), have not yet been reported for C. reinhardtii.
In this work, we investigated the putrescine metabolism of C. reinhardtii by functional gene analysis and supplementation experiments, which served as a starting point for recombinant putrescine overproduction. Various metabolic engineering strategies were employed to achieve robust yields, including overexpression and characterization of key pathway enzymes as well as utilizing CRISPR/Cas9‐based genome editing for pathway modulation. Putrescine accumulation was maximized by combining genetic engineering with phototrophic high cell density cultivation.
Results and Discussion
Exploring the native C. reinhardtii putrescine metabolism
Chlamydomonas reinhardtii is a facultative photoautotroph and can utilize several environmental organic molecules to accelerate growth, for example, acetate (Sager and Granick, 1953), urea (Kirk and Kirk, 1978), amino acids (Calatrava et al., 2019) or cellulose (Blifernez‐Klassen et al., 2012), either by direct cellular import or after extracellular enzymatic degradation. Supplemented putrescine was shown to be internalized by C. reinhardtii (Theiss et al., 2004), but the uptake mechanism and bio‐availability remain unclear.
To examine the utilization of extracellular putrescine and to evaluate potential toxic effects, a mixotrophic culture was spiked with different concentrations spanning four orders of magnitude (0.001–10 mm, Figure 1b and S1). Low levels of putrescine (0.001–0.1 mm) did not affect growth rates (Figure S1C), whereas higher concentrations (above 1 mm) induced a moderate, dose‐dependent reduction in growth rates (GR value below 0.9). About 10 mm putrescine lowered the GR value to 0.82 and reduced final cell concentrations by 40%. This growth arrest was accompanied by significantly increased average cellular diameters for concentrations above 0.1 mm putrescine (Figure S1D), with an up to 30% increase in diameter at 10 mm putrescine. This slightly attenuated over the course of the cultivation. Both effects were markedly less pronounced compared to a similar setup employing non‐native cadaverine (Freudenberg et al., 2021), indicating a relatively high tolerance of environmental putrescine in C. reinhardtii.
External putrescine concentrations positively correlated with cellular titres (Figure S1B), but most putrescine could be recovered after cultivation. With reference to the untreated control, only small amounts of the supplemented putrescine (4–26 mg/L, up to 9%) were lost due to metabolism or degradation (Figure S1A). Interestingly, regardless of the applied concentration, extracellular putrescine did not sustain growth when C. reinhardtii strain UVM4 was cultivated under nitrogen‐depleted conditions (Figure S1E). Merely the cell size was increased up to 20% after day 3 for the higher dosage points (above 1 mm), which might be due to osmotic effects. These results suggest that uptake or extracellular degradation of putrescine is limited and that higher amounts cannot serve as relevant nitrogen source under these conditions. It is likely that supplemented putrescine instead adheres to the cell surface, especially when considering that intracellular putrescine can be recycled by amine oxidases.
Ornithine decarboxylase (ODC) catalyses the decarboxylation reaction to produce putrescine from ornithine, which is also an intermediate of arginine anabolism found in the chloroplast. Chlamydomonas reinhardtii harbours two native ODC genes, which were disrupted by CRISPR/Cas9‐based genome editing to elucidate their individual contribution to putrescine accumulation. For ODC1 and ODC2, an editing frequency of 43% ± 1.5% (n = 2) and 75% ± 6% (n = 2) was achieved in pre‐selected transformants. Representative colony PCR (cPCR) results are found in Figure S2. Identified ODC knockout mutants (ΔODC) were cultivated mixotrophically and cellular putrescine levels in ΔODC1 strains were found to be significantly reduced by 66% (from 0.12 to 0.04 pg/cell, Figure 1c), whilst total volumetric yields were decreased by 77% (from 3.9 to 0.9 mg/L, Figure S3B). This highlights the importance of ODC1 as the main constituent for native putrescine synthesis, but also implies that additional enzymes are involved, as putrescine was not eliminated in ΔODC1 mutants. Disruption of ODC2 did not significantly alter putrescine titres (total and cellular) or cell growth (Figure 1c and S3C). We hypothesize that both ODC enzymes are independently regulated, whereby ODC1 activity is likely increased upon knockout of ODC2 to achieve putrescine homeostasis. On the other hand, ODC2 can only partially complement ODC1 activity, possibly because its expression does not involve a similar alternative splicing mechanism, but responds to nitric oxide (NO) instead (Zalutskaya et al., 2020). Several attempts at combining both ODC knockouts in a single strain were unsuccessful, even under supplemented polyamine conditions (5 μm spermidine and 0.1 mm putrescine, according to Freudenberg et al., (2022)). This suggests that alternative pathways (e.g. the ADC route) do not provide sufficient putrescine amounts in C. reinhardtii for survival.
Growth rates and final cell concentrations of ΔODC1 strains were reduced, whilst average cell diameters increased by up to 20% compared to the parental strain (Figure S3A). If putrescine was exclusively required as a precursor for synthesis of higher polyamines, adding spermidine should alleviate the negative growth effect of low putrescine levels. However, when supplementing 5 μm spermidine, growth and cell sizes were unchanged. The addition of 0.1 mm putrescine, on the other hand, resulted in slightly increased growth rates and a significant reduction in cell size comparable to that of the control strain N‐UVM4 (Figure S3A). It appears that putrescine directly fulfils important cellular functions required for cell proliferation. In particular, DNA and RNA stabilization or the synthesis of norspermidine might be affected, although the route of synthesis has not been elucidated yet (Tassoni et al., 2018). Interestingly, upon the addition of the ODC inhibitor α‐difluoromethylornithine (DFMO), cell numbers and cellular norspermidine content increased, whereas putrescine accumulation and ODC activity decreased (Tassoni et al., 2018). The possibility that DFMO acts as an abiotic stress agent in addition to its inhibitory effect on ODC was discussed by Tassoni et al. and is supported by our results since the knockout of ODC1 not only led to a decrease in cellular putrescine but also reduced growth rate and final cell concentrations.
Manipulating putrescine accumulation by overexpression of ornithine decarboxylases
Elevated putrescine accumulation was established by engineered expression of four different ODCs in C. reinhardtii strain N‐UVM4: The active splice variant of native C. reinhardtii ODC1 (CrODC1‐S), a plant ODC from Atropa belladonna (AbODC), which was previously utilized for tropane alkaloid production (Zhao et al., 2020) and two bacterial ODCs from Escherichia coli (EcoSpeC) and Enterobacter cloacae (EcSpeF), which have been employed for bacterial putrescine production (Li et al., 2018; Schneider and Wendisch, 2010).
The optimal construct design was tested for bacterial EcSpeF by comparing the effects of chloroplast or cytosolic enzyme localization to determine subcellular substrate availability (Lauersen et al., 2016) and C‐ or N‐terminal fluorescence reporter fusion (Figure S4). According to Western blotting results, chloroplast‐targeted expression was limited compared to cytosolic localization and no enzymatic activity, reflected by unchanged putrescine levels and could be observed. In addition, no significant difference was found in the placement of the fluorescence reporter. Our results suggest that ornithine is freely available in the cytosol of C. reinhardtii, reporter fusion does not inhibit enzyme activity and enzyme accumulation is more stable in the cytosol.
Candidate ODCs were cloned into suitable MoClo devices (Figure 2a) and used for nuclear transformation of C. reinhardtii strain N‐UVM4. Isolated transformants were characterized regarding cell growth, reporter fluorescence and volumetric putrescine yield from the cellular fraction (Figure 2b). Overexpression of CrODC1‐S (A1) had little effect on cell concentrations, resulted in very low reporter fluorescence and a broad range of putrescine titres ranging from 4 mg/L (native levels) to more than 14 mg/L. Expression of AbODC resulted in the highest average volumetric yields (ranging from 8 to 15 mg/L), which correlated with respective fluorescence intensities. Accordingly, construct A2 was used in the following engineering attempts. In line with the previous experiments on putrescine toxicity (Figure S1), a fourfold increase in cellular putrescine levels in case of AbODC expression (Figure S5) led to slightly reduced cell concentrations. It is possible that intracellular putrescine titres are much higher upon ODC overexpression compared to extracellular treatment with putrescine. However, physiological responses are comparable.
Figure 2.
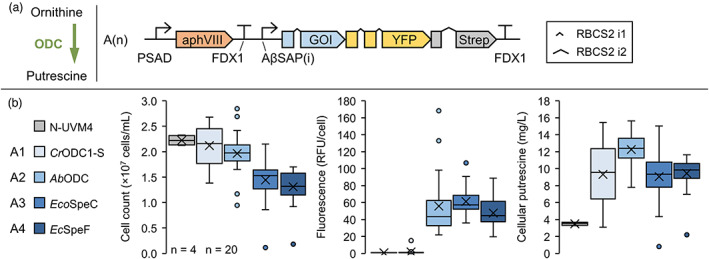
Establishing recombinant putrescine production in Chlamydomonas reinhardtii by recombinant expression of ornithine decarboxylases. (a) Decarboxylation reaction from ornithine to putrescine (left) and transgene design used in this experiment (right). (b) Strains with highest reporter fluorescence levels were cultivated mixotrophically for 4 days and compared to the parental strain N‐UVM4. The box‐and‐whisker plots depict the final cell concentration on day 4, adjusted YFP fluorescence level per cell and the volumetric cellular putrescine yield (from left to right) depending on the gene of interest (GOI). The number of biological replicates is indicated at the bottom left (n = 4 for N‐UVM4, n = 20 for A1‐A4). The number of introns in the GOI from A1 to A4 equals 4, 3, 5 and 5. Genetic symbols are based on the SBOL 3.0 suggestions or have been published previously (Crozet et al., 2018). The glyphs for promoter and terminator do include 5′ and 3′ UTRs, respectively. RFU, relative fluorescence units; PSAD Photosystem I reaction centre subunit II; FDX1, Ferredoxin; RBCS2 i1/i2, Ribulose bisphosphate carboxylase small subunit intron 1/2; AβSAP(i), HSP70A/βTUB2 Synthetic Algae Promoter; aphVIII, Aminoglycoside 3′‐phosphotransferase type VIII. [Colour figure can be viewed at wileyonlinelibrary.com]
Expression of both bacterial ODCs resulted in severely reduced final cell concentrations, despite only moderately elevated volumetric putrescine yields. We uncovered that both enzymes led to the formation of the two secondary products cadaverine and 4‐aminobutanol (Figure S5), which were identified via HPLC (Figure S6B,C) and UHPLC–MS/MS (Figure S7), respectively. The formation of 4‐aminobutanol has not yet been reported as a side product from bacterial ODCs and both compounds were not found in cells expressing AbODC, indicating they did not result from putrescine degradation. Substrate promiscuity of ornithine decarboxylases in some bacteria and plants has been observed under certain conditions (Guirard and Snell, 1980; Lee and Cho, 2001; Sakai et al., 1997; Takatsuka et al., 1999) and likely originates from structural similarities to arginine or lysine decarboxylases as they share a common ancestor within group IV of PLP (pyridoxal phosphate)‐dependent decarboxylases (Michael, 2016b).
Production of 4‐aminobutanol was evaluated further in a phototrophic, high cell density cultivation (6xP medium, Figure S8). It was found that the expression of both enzymes, EcoSpeC and EcSpeF, resulted in a severe reduction of final biomass and cell concentrations (on average 49.5% and 66%, respectively). A mean volumetric 4‐aminobutanol production of 66 mg/L (80 mg/L maximum) was achieved. In addition, about 40 mg/L of putrescine accumulated and cadaverine levels differed significantly between the two transformant populations, accumulating to 30 mg/L (EcoSpeC) and 13 mg/L (EcSpeF), respectively. Interestingly, putrescine and cadaverine were located primarily in the cellular fraction (80% and 70%, respectively), whereas 4‐aminobutanol was found in the supernatant (90%), suggesting that it was actively exported from the cell. Recently, the first bio‐production of 4‐aminobutanol was shown in engineered Corynebacterium glutamicum, achieving up to 24.7 g/L in a fed‐batch cultivation (Prabowo et al., 2020).
A potential candidate substrate in C. reinhardtii for bacterial ODCs to yield 4‐aminobutanol by decarboxylation is L‐pentahomoserine (5‐hydroxy‐L‐norvaline, CAS 6152‐89‐2). It was found in bacteria (Hill et al., 1993) and plants (Carmo‐Silva et al., 2009) before, but its existence in C. reinhardtii is unclear. Since it represents the reduced form of the proline biosynthesis intermediate glutamate‐5‐semialdehyde (Figure 1a), some species (e.g. Neurospora crassa) are able to utilize it for protein biosynthesis (Vogel and Kopac, 1959). Another possibility is that the two bacterial ODCs exhibit a latent ability to catalyse a decarboxylation‐dependent oxidative deamination of ornithine to yield 4‐aminobutanal, which could be reduced by C. reinhardtii to form 4‐aminobutanol (Bertoldi et al., 1999; Pontrelli et al., 2018). Enzyme promiscuity was also observed when expressing a bacterial lysine decarboxylase in C. reinhardtii (Freudenberg et al., 2021), which, under certain conditions, led to an abundant side product accumulation in the supernatant of a high cell density cultivation. These results demonstrate limitations in the application of recombinant enzymes from distantly related species and emphasize the need for systematic characterization of candidate enzymes in heterologous hosts.
Putrescine production correlates with growth but is reduced by substrate supplementation
Putrescine accumulation was evaluated during a mixotrophic cultivation of engineered N‐UVM4 strains expressing AbODC‐YFP (vector A2, Figure 3a). From day 2 onwards, cell concentrations were significantly reduced compared to the parental strain. Reduced cell viability was observed for all strains after day 4, likely due to elevated pH (around 9) after acetate consumption in TAP‐NO3. Due to this ongoing cell lysis, putrescine was found increasingly in the supernatant fraction at later cultivation time points, which was more pronounced in transformant strains, where 50% of total putrescine was found in the supernatant after 6 days. Furthermore, total putrescine accumulated rapidly during the initial growth phase and titres stabilized after day 2 around 17 mg/L.
Figure 3.
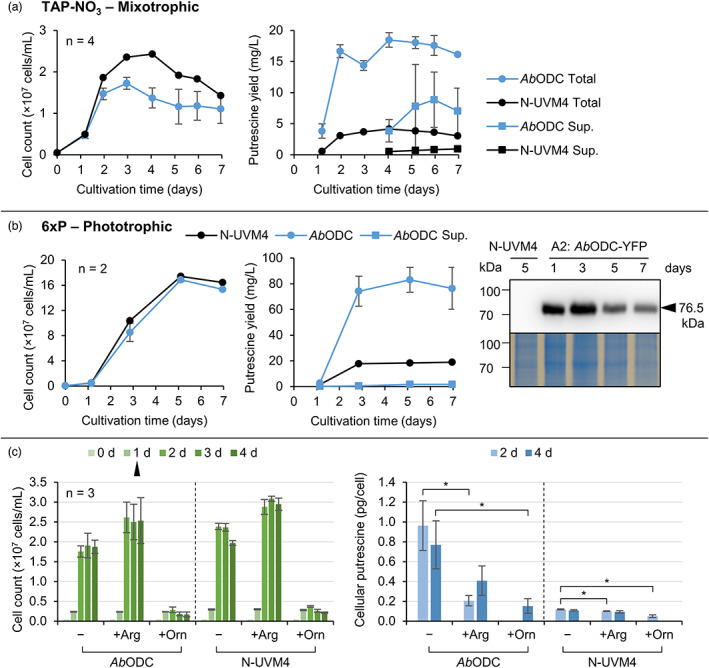
Evaluating recombinant putrescine production over time depending on the cultivation regime and medium supplementation. (a) Mixotrophic cultivation of the strain N‐UVM4 (n = 1) and different strains exhibiting high reporter fluorescence levels after transformation with construct A2 (AbODC‐YFP) (n = 4). Cell concentrations are depicted on the left and putrescine yields on the right, where total putrescine and supernatant (Sup.) yields are plotted separately. The supernatant fraction was quantified beginning with day 4 and likely contained some putrescine on day 3. (b) Phototrophic, high cell density cultivation of the same strains as in (a) with cell concentrations (left), putrescine yields (middle) and expression levels over time of the AbODC‐YFP fusion protein visualized via Western blotting and detection using an anti‐Strep antibody (right). Error bars represent the standard deviation of biological replicates. (c) Cell concentrations (left) and cellular putrescine yield (right) during a mixotrophic cultivation of the same strains as in (a), which were spiked with 1 mM of either arginine (Arg) or ornithine (Orn) on day one (black triangle). Missing data points indicate results below the detection limit. Asterisks (*) indicate the significance level of an unpaired, two‐sided Student's t‐test assuming non‐homogenous variances (*P < 0.05). [Colour figure can be viewed at wileyonlinelibrary.com]
Since recombinant putrescine accumulation correlated with active cell division, prolonging the growth phase by cultivating the transformant strains phototrophically in high cell density medium (6xP) should increase titres. In addition to an almost 10‐fold increase in cell concentrations (Figure 3b), 83 mg/L putrescine accumulated, with only small amounts found in the supernatant, which implies that putrescine was not actively exported by the cells. No pH‐related reduction in cell concentrations of both the control and production strains occurred in the stationary phase. It can be concluded that unless cell lysis releases putrescine to the supernatant, very little transport or diffusion of putrescine occurs, which reflects the observations made with external putrescine (Figure S1). Interestingly, almost no cytostatic effects were observed in the production strain. This is explained by the relatively low cellular putrescine titres after day 3, which dropped below 0.43 pg/cell. It seems that carbon and nitrogen partitioning was shifted towards biomass rather than putrescine accumulation under these conditions, even though volumetric putrescine yield was higher than during mixotrophic growth and high recombinant gene expression (according to the Western blotting result) was detectable throughout the cultivation. A potential lack of substrate availability or increasing putrescine degradation might have caused the stagnant volumetric and decreasing cellular yields after day 3.
To increase substrate supply, a mixotrophic cultivation was complemented with additional 1 mm ornithine or arginine (Figure 3c). The presence of external ornithine completely abolished cell division in both the production and control strains, which is reflected by significantly lower biomass accumulation and total putrescine yields (Figure S9A). Further, ornithine was fully recovered from the culture, indicating that it was not metabolized, even though cellular import might have occurred (Figure S9B). In contrast, C. reinhardtii wild‐type strains were shown to grow on a solid medium supplied with up to 100 mg/L (0.76 mm) ornithine (Loppes, 1970), whilst at higher concentration (500 mg/L), a severe growth inhibition was reported (Sussenbach and Strijkert, 1969). The authors postulated that this observed growth arrest was due to blocked arginine utilization for protein biosynthesis via forward inhibition of arginyl‐tRNA synthetase by argininosuccinate. It is likely that a cell wall‐reduced strain such as UVM4 (Neupert et al., 2009) is much more susceptible to external ornithine, especially in liquid cultivations, where the mass transfer is faster.
Supplemented arginine was clearly metabolized by the cells (up to 80% after 4 days, Figure S9B) and resulted in increased final cell concentrations (Figure 3c) and biomass accumulation (Figure S9A). Arginine has been shown to serve as a direct (via carrier‐mediated import) and indirect (via enzymatic degradation releasing ammonia) nitrogen source for growth in C. reinhardtii (Calatrava et al., 2019; Kirk and Kirk, 1978). However, putrescine yields diminished by about 50% in production strains and were slightly reduced in the control strain, even though nitrogen was continuously available as both nitrate and arginine. Low ornithine titres might be responsible, whereby the internalized arginine reached the inhibition concentration of NAGK, shutting down the arginine anabolism pathway (Selim et al., 2020b). Feeding arginine as the sole nitrogen source is known to trigger an N starvation‐like response in C. reinhardtii (Munz et al., 2020), which could also lead to lower putrescine amounts by upregulation of degrading enzymes. To conclude, substrate availability might pose a limitation during ODC overexpression but cannot be circumvented by supplementation, due to the extensive regulation of arginine and putrescine biosynthesis.
Putrescine accumulation depends partially on ornithine decarboxylase enzyme titre
Increasing the cellular enzyme titres by multiple transformations of the same GOI is a common strategy for improving metabolic flux towards the desired product (Wichmann et al., 2018). Accordingly, an additional vector was designed (A5: AbODC‐RFP, Figure 4a) and used for transformation. The resulting strains (2xAbODC) were characterized in terms of cell concentrations and cellular putrescine titres (Figure 4b). Elevated ODC levels significantly reduced final cell concentrations by about 18% to 1.63 × 107 cells/mL and increased cellular putrescine yields during exponential and stationary growth phase by 12% and 19% (to 0.82 and 0.68 pg/cell), respectively. Both effects resulted in a non‐significant change in volumetric yields and in continued cellular putrescine loss over time. It is likely that reduced cell fitness due to product accumulation or substrate limitation inhibits further increase in putrescine accumulation.
Figure 4.
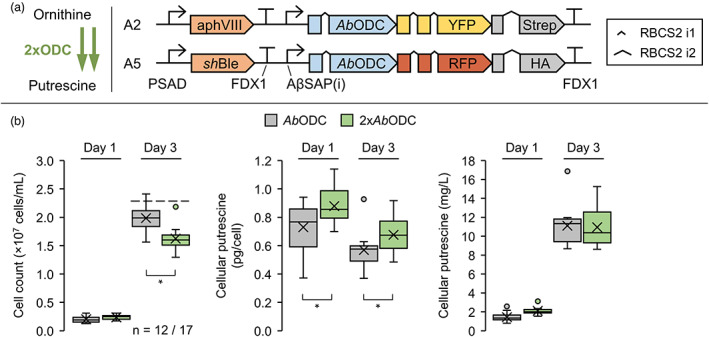
Effect of gene stacking on recombinant putrescine production. (a) Desired increase in the decarboxylation reaction from ornithine to putrescine (left) and design of the transgene constructs used in this experiment (right). (b) Strain N‐UVM4 was transformed with either construct A2 (AbODC‐YFP) or A5 (AbODC‐RFP) and subsequently with construct A5 or A2, respectively, to develop strains that expressed two copies of the AbODC GOI. The strains with highest reporter fluorescence (n = 17) were cultivated alongside their parental strains (n = 12) mixotrophically for 3 days and the resulting cell concentrations and putrescine yields extracted from the pellet fraction are depicted in the box‐and‐whisker plots (left to right) for days one and three. The dashed line indicates the mean cell concentration of the N‐UVM4 strain. Asterisks (*) indicate the significance level of an unpaired, two‐sided Student's t‐test assuming non‐homogenous variances (*P < 0.05). Genetic symbols are based on the SBOL 3.0 suggestions or have been published previously (Crozet et al., 2018). The glyphs for promoter and terminator do include 5′ and 3′ UTRs, respectively. Promoters and terminators are the same for constructs A2 and A5. PSAD, Photosystem I reaction centre subunit II; FDX1, Ferredoxin; RBCS2 i1/i2, Ribulose bisphosphate carboxylase small subunit intron 1/2; AβSAP(i), HSP70A/βTUB2 Synthetic Algae Promoter; aphVIII, Aminoglycoside 3′‐phosphotransferase type VIII; shBle, Streptoalloteichus hindustanus Ble protein. [Colour figure can be viewed at wileyonlinelibrary.com]
Heterologous arginase expression for increased substrate provisioning
Robust substrate availability is crucial for efficient ODC productivity. In addition to biosynthesis from glutamate, cellular ornithine can be provided by a heterologous arginase, which catalyses the hydrolytic reaction of arginine to ornithine and urea (Figure 5a). Whilst urea serves as an additional nitrogen source, reduced arginine levels should stimulate the polyamine biosynthesis pathway. In addition, supplementation of high ornithine concentrations and resulting growth arrest can be avoided.
Figure 5.
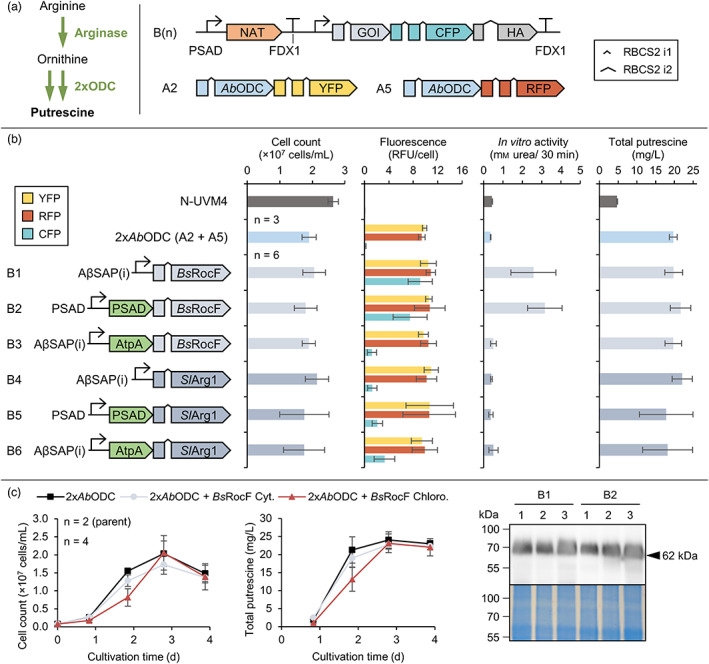
Improving ornithine substrate availability by expression of recombinant arginases. (a) Arginase reaction leading to a desired increase in ornithine for subsequent decarboxylation by recombinant ODCs (left) and design of the transgene constructs used in this experiment (right). Expression elements for constructs A2 and A5 are omitted for clarity (see Figure 4). (b) Strains expressing two copies of the AbODC GOI (constructs A2 and A5) with highest reporter fluorescence levels were transformed with constructs B1‐6 and cultivated mixotrophically for 3 days. The charts depict cell concentrations, adjusted reporter fluorescence, in vitro arginase activity (quantified as mm urea produced in 30 min) as well as total putrescine yields (left to right) depending on the genetic construct. (c) Strains expressing ODCs from vectors A2 (AbODC‐YFP) and A5 (AbODC‐RFP) as well as B1 (BsRocF‐CFP) or B2 (BsRocF‐CFP targeted to the chloroplast) were cultivated mixotrophically for 4 days. Cell concentrations and total putrescine yields are depicted (left) together with visualization of the BsRocF‐CFP fusion protein by Western blotting using an anti‐HA antibody for detection (right) for three different N‐UVM4 transformants. Genetic symbols are based on the SBOL 3.0 suggestions or have been published previously (Crozet et al., 2018). The glyphs for promoter and terminator do include 5′ and 3′ UTRs, respectively. The number of introns in the GOI for BsRocF and SlArg1 equals 3. The PSAD chloroplast and AtpA mitochondrial targeting peptides are shown in green. Error bars represent the standard deviation of biological replicates. RFU, relative fluorescence units; PSAD, Photosystem I reaction centre subunit II; FDX1, Ferredoxin; RBCS2 i1/i2, Ribulose bisphosphate carboxylase small subunit intron 1/2; AβSAP(i), HSP70A/βTUB2 Synthetic Algae Promoter; NAT, Nourseothricin N‐acetyl transferase. [Colour figure can be viewed at wileyonlinelibrary.com]
In plants, arginases are localized in the cytoplasm or mitochondria and are responsible for the mobilization of stored nitrogen as arginine and the generation of ornithine as stress response (Siddappa and Marathe, 2020). Intriguingly, no native arginase can be found in the Chlamydomonas genome, which reduces the probability of inhibition by regulatory elements.
Arginase coding sequences were chosen from Bacillus subtilis (BsRocF) and from the plant Solanum lycopersicum (SlArg1), as both have been characterized before (Chen et al., 2004; Yu et al., 2013). Only manganese is required as cofactor, which is supplied by the TAP medium (Kropat et al., 2011). Six expression vectors were constructed to facilitate targeted expression of the two arginases to all relevant compartments (Figure 5a,b). After transformation of the putrescine production strain (harbouring two copies of the AbODC expression cassette), a systematic characterization based on initial colony fluorescence was conducted by determining cell concentrations, reporter fluorescence, in vitro arginase activity and total putrescine yields. No significant effect on final cell concentration was observed and reporter fluorescence of the two ODC cassettes (A2 and A5) indicated consistently high expression. Arginase reporter fluorescence, however, was low (below 36% of maximal values) for all vectors except B1 and B2 (BsRocF‐CFP located in the cytosol or chloroplast). This indicates poor expression capacity of SlArg1‐CFP and impaired accumulation of BsRocF‐CFP in the mitochondria. Ornithine titres remained below the detection limit in all strains, necessitating an alternative quantification method. In vitro arginase activity, which was quantified directly after whole cell lysis, correlated well with reporter fluorescence. Only cells derived from transformations with vectors B1 and B2 exhibited appreciable in vitro activity above control levels (0.4 mm urea) with about 3 mm urea (6.5‐fold increase) formed after 30 min. Protein characterization in N‐UVM4 demonstrated that the fusion protein BsRocF‐CFP was functionally expressed (Figure 5c); however, total putrescine yields were not increased for both putrescine production lineages expressing arginases of vectors B1 and B2. This was verified by a time‐resolved production experiment (Figure 5c). With strains expressing BsRocF‐CFP (targeted to the chloroplast), cell concentrations were markedly lower on day 2, but no other difference to the parental strains was observed.
Although our results confirm in vitro activity and intact enzyme accumulation, no significant increase in putrescine titres could be observed. It is likely that either product inhibition by putrescine limited ODC activity that ODC substrate limitation was not a bottleneck in this engineering or that elevated putrescine degradation counteracted any increase in production. Additionally, it is conceivable that the cell volume restricts further putrescine accumulation.
AMX2 is a putrescine degrading enzyme
In C. reinhardtii, endogenous putrescine serves as substrate for two enzymatic routes (Figures 1a and 6a) involving (i) the copper‐containing amine oxidases AMX1‐3 and (ii) spermidine synthase SPD1 (Harris et al., 2009). Previous work revealed that functional SPD1 expression is essential for cell proliferation (Freudenberg et al., 2022), but spermidine levels are comparably low, suggesting little metabolic flux via this route.
Figure 6.
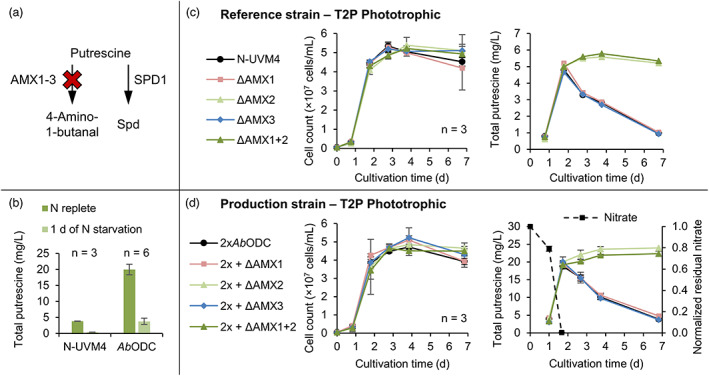
Engineering the putrescine catabolism in Chlamydomonas reinhardtii by CRISPR/Cas9‐mediated knockout of key enzymes. (a) Relevant reactions of putrescine utilization and degradation. The latter is highlighted by a red cross. (b) Effect of nitrogen starvation on total putrescine yields in strain N‐UVM4 and transformants expressing AbODC‐YFP (vector A2). On day 3 of a mixotrophic cultivation, cells were transferred to nitrogen depleted TAP medium and cultivated for another day. (c) Utilizing CRISPR/Cas9, the genes AMX1‐3 were targeted and knocked out in the reference strain N‐UVM4 by insertion of an antibiotic resistance cassette. Dual knockouts of the genes AMX1 and AMX2 were created by consecutive transformations. Strains were cultivated phototrophically and cell concentrations and total putrescine yields are plotted. (d) Similar to (c), the genes AMX1‐3 were targeted and knocked out in the putrescine production strain, expressing AbODC‐YFP and AbODC‐RFP (vectors A2 and A5). Dual knockouts of the genes AMX1 and AMX2 were created by consecutive transformations. Strains were cultivated phototrophically and in addition to cell concentrations and total putrescine yields, the remaining nitrate in the culture supernatant is plotted. Error bars represent the standard deviation of biological replicates. AMX, Amine oxidase; SPD, Spermidine synthase; Spd, Spermidine. [Colour figure can be viewed at wileyonlinelibrary.com]
However, the three amine oxidases AMX1‐3 are promising candidates for further engineering, as they have been intimately associated with nitrogen metabolism and putrescine degradation to 4‐aminobutanal, H2O2 and ammonia. AMX1 and AMX2 share 47.1% amino acid sequence identity, whereas AMX3 only shares 22.8% identity with AMX1 and 21.5% with AMX2. Expression of all three genes is strongly induced upon nitrogen deprivation (log2‐fold change of AMX1: 8–10, AMX2: 3.5–7, AMX3: 5.8–9.3), but the absolute expression of AMX3 is comparably low (about eightfold less) in all tested conditions (Schmollinger et al., 2014). The effects of nitrogen starvation on the putrescine yield during a mixotrophic cultivation are shown in Figure 6b. In line with AMX expression data, a reduction of 89% (from 3.8 to 0.41 mg/L) and 81% (from 20 to 3.8 mg/L) in total putrescine titres could be observed after 1 day of cultivation in both the control strain and the putrescine production strains, respectively.
CRISPR/Cas9‐based genome editing was used to functionally inactivate AMX1‐3 in the control strain N‐UVM4 and the production strain (harbouring two copies of the AbODC cassette, A2 and A5). Comparable high editing frequencies according to cPCR after pre‐selection were achieved: AMX1: 67% ± 18% (n = 4), AMX2: 78% ± 16% (n = 2), AMX3: 54% ± 18% (n = 2). Representative cPCR results are shown in Figure S2 and exemplary sequencing data for ΔAMX1 are depicted in Figure S10. In addition, a subsequent knockout of AMX2 was introduced into ΔAMX1 mutant strains to unravel potential cumulative effects (ΔAMX1 + 2). Three independent mutant strains for each gene knockout were cultivated phototrophically in T2P‐NO3 medium (Figure 6c,d), where nitrogen limitation was reached after 2 days, as nitrate levels were drastically reduced (Figure 6d). No significant differences in growth characteristics were observable. However, whilst total putrescine titres in all N‐UVM4 strains increased to about 5 mg/L until day 2 and dropped to 1 mg/L after the onset of nitrogen starvation, yields remained stable till the end of the cultivation in N‐UVM4 ΔAMX2 and ΔAMX1 + 2 strains. No cumulative effect of the double knockout of AMX1 and AMX2 was discernable, pointing to AMX2 as the main enzyme involved in putrescine degradation. A similar result was obtained from AMX knockout mutants derived from the production strain (Figure 6d), where growth was also unaffected, but generally increased putrescine levels (20 mg/L after 2 days) were obtained. Production yields remained stable in 2xAbODC ΔAMX1 and ΔAMX1 + 2 strains, whereas they were reduced substantially upon nitrogen limitation in all other strains.
When cultivating the production strain and associated AMX knockout mutants mixotrophically in a TAP medium, where nitrogen limitation occurs much later, no significant effect of the AMX2 knockout on putrescine accumulation was observed (Figure S11). Apparently, no putrescine degradation was occurring via the AMX route in these conditions, indicating that AMX2 expression is tightly regulated and associated with nitrogen limitation.
We conclude that AMX2 is the main contributor to putrescine oxidation during nitrogen limitation. Although AMX1 and AMX3 are co‐expressed under the same condition, their roles remain elusive, as a functional knockout had no effect on putrescine levels. It is very likely that these enzymes act on alternative substrates (e.g. amino acids) and AMX2 is the only enzyme capable of putrescine oxidation during nitrogen limitation.
Maximizing putrescine accumulation by combining high cell density cultivation with metabolic engineering
Phototrophic, high cell density cultivation is a powerful strategy to increase volumetric productivity of all recombinant products in C. reinhardtii and can be achieved by adjusted nutrient salt contents and high light and CO2 supply (Freudenberg et al., 2021). Candidate strains from the three engineering attempts (i) 2xAbODC; (ii) 2xAbODC + BsRocF and (iii) 2xAbODC ΔAMX2 were compared to the control strain N‐UVM4 utilizing the CellDEG HD100 cultivators (Figure 7a). CO2‐enriched air (10% v/v) and gradually increased light intensities from 450 to 1000 μmol photons/m2/s resulted in robust growth and biomass accumulation. Detailed performance analysis (Figure 7b) revealed that nitrate and phosphate were removed from the supernatant by day 7 of the cultivation, at which time, the effects of the AMX2 knockout manifested: Total putrescine yields in ΔAMX2 strains were significantly higher than in the other strains. Interestingly, no severe degradation of putrescine was observed, suggesting that nitrogen was not a limiting factor for growth during the stationary phase. Light stress, self‐shading or deprivation of a nutrient salt besides nitrate might have limited growth. We hypothesize that AMX2 is likely not induced under these conditions.
Figure 7.
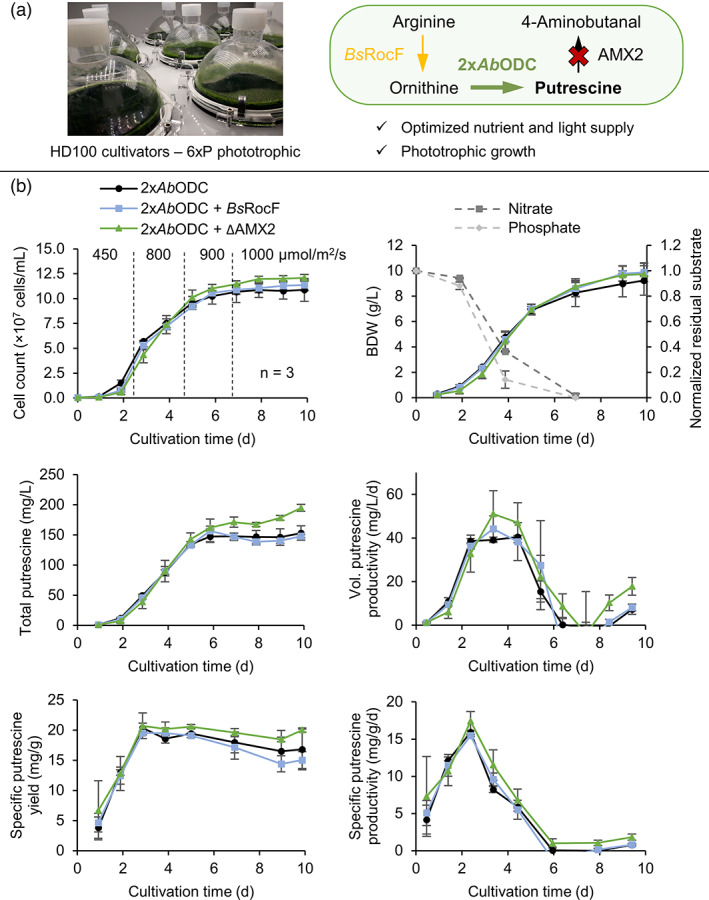
Phototrophic, high cell density cultivation of the best putrescine production strains obtained during the previous investigations in Figures 5 and 6 utilizing the CellDEG HD100 cultivators. (a) Picture of the HD100 cultivators and summary of the most important engineering strategies for increased putrescine accumulation. Overexpression of a high‐performance ODC enzyme was coupled with knockout of the native degrading enzyme AMX2 and optimized nutrient supply in a phototrophic cultivation. Improved substrate provisioning by arginase expression might have an effect in phototrophic conditions. (b) Detailed performance parameters of the putrescine production strains and incident light intensities. Cell concentrations, biomass dry weight (BDW), average residual nitrate and phosphate in the supernatant, as well as volumetric and specific putrescine yields and productivities are plotted. Error bars represent the standard deviation of biological triplicates. [Colour figure can be viewed at wileyonlinelibrary.com]
The best production strain achieved a maximal productivity of 58 mg/L/d (volumetric), 18.9 mg/g/d (specific) and a fairly stable specific yield of 20.5 mg/g BDW. The apparent rise in productivities from day 8 is due to increased medium evaporation under very high light conditions, which amounted to about 14.5% (of initial volume) on day 10.
Final volumetric putrescine yields of (i) 153 ± 12 mg/L; (ii) 148 ± 3 mg/L; and (iii) 195 ± 6 mg/L were achieved. The knockout of AMX2 combined with overexpression of two copies of AbODC resulted in highest volumetric putrescine yields of up to 200 mg/L. Expression of BsRocF did not significantly improve yields, indicating that ornithine substrate limitation was unlikely. In summary, through state‐of‐the‐art metabolic engineering strategies, putrescine levels were raised 10‐fold relative to the control strain N‐UVM4 from 20 mg/L in these optimized growth conditions (see Figure S12 for performance data of N‐UVM4).
This study demonstrates the first phototrophic bio‐production of putrescine. Previous heterotrophic fermentations yielded 6 g/L putrescine (batch cultivation) with up to 0.1 g/L/h in engineered C. glutamicum (Schneider and Wendisch, 2010). Later, 12.5 g/L (batch) was achieved and fed‐batch attempts with engineered Escherichia coli resulted in up to 42.3 g/L (Li et al., 2018). However, when grown on more sustainable, pre‐treated rice straw hydrolysate as carbon source yields were considerably lower with 91 mg/L after 3 days (Sasikumar et al., 2021). This is comparable to the achievements presented here and makes C. reinhardtii an effective competitor when it comes to sustainable, phototrophic bio‐production of polyamines.
Conclusions and Outlook
Despite its native abundance in C. reinhardtii, engineered putrescine bio‐production is comparably challenging due to several present layers of regulations. Biosynthesis is influenced by adaptive metabolic response to available nitrogen, carbon and metabolites such as ornithine and arginine. Our results suggest that putrescine accumulation is closely linked to nitrogen availability and is rapidly mobilized upon nitrogen deprivation. Even though cells can tolerate high external putrescine concentrations (<0.44 g/L), import and utilization was limited and unable to compensate a lacking nitrogen source. By state‐of‐the‐art CRISPR/Cas9‐based gene knockouts, we confirmed that ODC1 is the main enzyme for putrescine homeostasis. ODC2 contributes up to a third of physiological titres in case of ODC1 knockout, whilst the ADC pathway is likely inactive. Furthermore, AMX2 is shown to solely mediate putrescine oxidation during nitrogen limitation, whilst AMX1 and AMX3 likely accept other substrates.
This study displays the first recombinant and phototrophic putrescine production in C. reinhardtii, achieved via several advanced metabolic engineering approaches. Successful knockout of AMX2 to prohibit product degradation was coupled with an effective overexpression of two copies of the robust plant AbODC. This resulted in the highest volumetric yields of 200 mg/L, when combined with an optimized, phototrophic high cell density cultivation strategy (Figure 7a). Ornithine was shown to be freely available in the cytosol of C. reinhardtii and titres could not be increased by expression of a recombinant arginase. We uncovered a yet unreported substrate promiscuity of bacterial ODCs, which when expressed in the cytosol of C. reinhardtii led to the formation of cadaverine and high amounts of 4‐aminobutanol. This compound was likely exported to the culture medium and poses a promising alternative target for bio‐production (Prabowo et al., 2020).
Putrescine production and ornithine availability would benefit from a raised glutamate synthesis. Here, a key regulating enzyme, the guanylate cyclase CYG56 is a promising candidate for further engineering to circumvent glutamate synthesis inhibition in excess arginine or ammonium conditions (González‐Ballester et al., 2018). Engineered glutamate acetylation via N‐acetylglutamate kinase (NAGK) and improved nutrient supply could further stimulate metabolic flux towards putrescine. Importantly, putrescine export to the culture supernatant should be explored to avoid product inhibition and reduce its effect on cellular fitness.
Methods
Vector design and cloning
Plasmid design was based on the standardized modular cloning system (MoClo) (Crozet et al., 2018) including recent design improvements (Einhaus et al., 2022). Coding sequences of CrODC1‐S (UniProt: A8J7E8), AbODC1 (A0A060IEJ3), EcoSpeC (P21169), EcSpeF (A0A0H3CLT7), BsRocF (P39138) and SlARG1 (Q5UNS2) were synthetically optimized for high transgene expression (Table S1) (Baier et al., 2018; Jaeger et al., 2019), followed by gene synthesis (GenScript Biotech B.V., Leiden, Netherlands). Note that CrODC1‐S is based on transcript t2.1 (Phytozome: Cre03.g159500) and that as of genome version 5.6 another, larger version might exist, which retains exon 4 in addition to a newly described short exon just upstream (transcript t1.2). However, experimental data are lacking and an earlier study identified transcript t2.1 as the only one leading to a functional enzyme (Pandey et al., 2020). The fluorescence reporters mVenus (YFP, NCBI: AAZ65844), mRuby2 (RFP: AFR60232) and mCerulean3 (CFP: ATP07149) were implemented as previously described (Baier et al., 2020) and translationally fused along with a terminal Strep II or HA epitope tag.
Transcription was driven by the HSP70A/βTUB2 Synthetic Algae Promoter (AβSAP(i)) (Einhaus et al., 2021)), HSP70A/RBCS2 (AR(i)) (Schroda et al., 2000) or photosystem I reaction centre subunit II (PSAD) promoter (Fischer and Rochaix, 2001) and the ferredoxin (FDX1) terminator (López‐Paz et al., 2017). Post‐translational transport to mitochondria or the chloroplast was initiated by incorporation of the signal peptides from the ATP synthase subunit alpha (AtpA) or PSAD, respectively. Antibiotic selection was facilitated by a separate module, expressing aminoglycoside phosphotransferase type VII, VIII (aphVII (Berthold et al., 2002), aphVIII (Sizova et al., 2001)), Streptoalloteichus hindustanus Bleomycin resistance protein (shble) (Stevens et al., 1996), aminoglycoside adenylyltransferase aadA (Cerutti et al., 1997) or nourseothricin N‐acetyl transferase (NAT) (Yang et al., 2019). The PSAD promoter and FDX1 terminator controlled expression of all antibiotic resistance modules.
Repair template (donor‐DNA) plasmids for CRISPR/Cas9 editing experiments were constructed as previously described (Freudenberg et al., 2022). Briefly, two fragments of genomic DNA (homology arms) of around 500 bp each up‐ and downstream of the sgRNA target area were amplified from the C. reinhardtii genome via PCR (Q5 High Fidelity polymerase, NEB). Primers included overhangs for direct level 1 module cloning via cut‐ligation (primers in Table S2) as previously described (Crozet et al., 2018). An antibiotic resistance module was included in the level 2 device assembly.
Assembled plasmids were used to transform chemically competent E. coli DH5α cells and selection was carried out on solid LB agar plates containing the appropriate antibiotics (level 0: 50 mg/L streptomycin, level 1: 300 mg/L ampicillin, level 2: 50 mg/L kanamycin). Plasmids were extracted using the peqGOLD plasmid isolation kit (VWR), followed by Sanger sequencing (Sequencing Core Facility, Bielefeld University).
Chlamydomonas reinhardtii cultivation
Chlamydomonas reinhardtii strains were maintained under mixotrophic growth conditions on solid Tris‐acetate phosphate (TAP) (Gorman and Levine, 1965) agar plates with updated trace element composition (Kropat et al., 2011). A continuous photon flux density of 50 μmol/m2/s was applied. Nitrogen was provided as either ammonium for strain UVM4 (Neupert et al., 2009) or nitrate for the nitrate‐capable variant N‐UVM4 (Freudenberg et al., 2021).
Liquid cultivations were inoculated with 5–7.5 × 105 cells/mL and carried out either mixotrophically in TAP medium and ambient air or phototrophically in T2P medium (Lauersen et al., 2016) with CO2‐enriched air (3–5% v/v) and a continuous photon flux density of around 350 μmol/m2/s. For high cell density cultivations, 6xP medium (Freudenberg et al., 2021) supplied with up to 10% (v/v) CO2‐enriched air was used in either 6‐well plates or the HD100 cultivators (CellDEG GmbH) with a working volume of 110 mL.
Cell concentration and average cellular diameter were quantified with the Z2 Coulter Counter Analyser (Beckman Coulter Life Sciences, Krefeld, Germany) following the manufacturer's instructions. Biomass dry weight (BDW) was determined gravimetrically after centrifugation (5 min at 20 000 g) of a 2 mL culture sample and drying the pellet overnight at 105 °C. Dissolved nitrate and ortho‐phosphate in the culture supernatant were quantified using the LCK 339 and LCK 349 (Hach Company, Düsseldorf, Germany) cuvette test kits together with the DR 3900 VIS spectrophotometer (Hach Company) according to the manufacturer's instructions.
Nuclear transformation and CRISPR/Cas9‐based genome editing
The nuclear transformation was performed via the glass bead method (Kindle, 1990) using 10 μg of linearized plasmid DNA and 3 × 108 cells in the exponential growth phase. Transformants were selected on TAP agar plates containing the appropriate nitrogen source and antibiotics (10 mg/L Paromomycin, 10 mg/L Hygromycin, 10 mg/L Zeocin, 2.5 mg/L Nourseothricin or 200 mg/L Spectinomycin), applying continuous light of around 500 μmol photons/m2/s.
Nuclear genome editing was achieved by direct delivery of pre‐assembled Cas9‐sgRNA ribonucleoproteins (RNPs) and double‐stranded donor‐DNA (repair template, containing homologous flanking regions) via electroporation as previously described (Freudenberg et al., 2022). Briefly, for each target gene, sgRNAs (Table S3) were determined with the online Cas‐Designer (RGEN tools) and synthesized using the EnGen® sgRNA Synthesis Kit (New England Biolabs GmbH, NEB) following the manufacturer's instructions. Target site digest capability was tested in vitro by incubating PCR‐amplified genomic DNA with pre‐assembled Cas9‐sgRNA RNPs for 30 min at 37 °C, followed by separation via agarose gel electrophoresis (Figure S13).
Chlamydomonas reinhardtii cells were transferred to nitrogen‐depleted TAP medium 24 h prior to electroporation and a total of 7 × 107 cells were harvested by centrifugation (1000 g for 3 min) and resuspension in 120 μL of TAP‐sucrose (40 mm), followed by heat shock treatment at 40 °C for 20 min. RNPs were assembled at room temperature (8 μg Cas9‐NLS (NEB), 7 μg sgRNA) and combined with 750 ng linearized repair template. Electroporation was conducted (Square‐wave protocol, 2 mm electrode gap, 250 V, single 8 ms pulse) with the Gene Pulser Xcell System (Bio‐Rad Laboratories GmbH, Feldkirchen, Germany) and cells were subsequently recovered at low light (10 μmol photons/m2/s) for 24 h, followed by selection on agar plates containing the appropriate nitrogen source and antibiotic at 300 μmol photons/m2/s.
Emerging colonies (at least 22) were screened for a successful edit of the target gene by colony PCR (cPCR) as described previously (Cao et al., 2009) using Q5 High Fidelity Polymerase (NEB) (primers in Table S2). PCR products were separated via agarose gel electrophoresis (Figure S2) and sequence identity was confirmed by Sanger sequencing (Sequencing Core Facility, Bielefeld University). Functional investigations were conducted in at least three independent knockout strains.
Fluorescence screening
Fluorescence intensity was quantified in liquid culture and on agar plates as previously described (Lauersen et al., 2016). Briefly, an initial pool of at least 144 colonies per transformation was isolated randomly and their fluorescence on plate level was compared via an in vivo plant imaging system (NightShade LB 985; Berthold Technologies GmbH & Co. KG, Bad Wildbad, Germany) with appropriate filters (YFP: ex. 504/10 nm, em. 530/20 nm, RFP: ex. 560/10 nm, em. 600/20 nm, CFP: ex. 425/40 nm, em. 500/20 nm). To reduce background fluorescence, TAP agar plates were supplemented with 250 mg/L amido black 10B (Carl Roth GmbH + Co. KG, Karlsruhe, Germany). Fluorescence levels of individual colonies were verified using a Leica M205 FA stereomicroscope (Leica Microsystems GmbH, Wetzlar, Germany) with appropriate filters (YFP: ex. 490–510 nm, em. 520–550 nm, RFP: ex. 540–580 nm, em. 589–599 nm, CFP: ex. 426–446 nm, em. 460–500 nm). Transformants with highest fluorescence intensities were cultured in microtitre plates and fluorescence was measured together with optical density at 750 nm in a plate reader (Infinite M200 PRO; Tecan Group Ltd., Männedorf, Switzerland) with appropriate filter settings (YFP: ex. 515/9 nm, em. 550/20 nm, RFP: ex. 560/9 nm, em. 590/20 nm, CFP: ex. 445/9 nm, em. 485/20 nm). Untransformed cells were used for signal normalization.
HPLC and polyamine analysis
Extraction and polyamine quantification were conducted for both, cellular and supernatant fractions as previously described (Freudenberg et al., 2021). Note that the term ‘cellular putrescine’ is used whenever the cellular fraction was exclusively extracted and used to calculate volumetric or per cell yields. To allow unambiguous detection of arginine via HPLC, TRIS buffer was replaced with HEPES buffer in respective experiments (McFadden and Melkonian, 1986). Briefly, a cultivation sample of 1 mL was centrifuged (3000 g for 3 min) and the cells were resuspended in 600 μL 5% (v/v) trichloroacetic acid (TCA) containing the internal standard diaminohexane (DAH, 5 mg/L; Acros Organics). Likewise, 0.9 mL of culture supernatant was combined with 0.1 mL 50% (v/v) TCA containing DAH (50 mg/L). After a 15 min incubation on ice, the resulting debris was pelleted (20 000 g for 8 min) and supernatants were used for pre‐column ortho‐phthalaldehyde (OPA) derivatization, followed by reversed‐phase HPLC and fluorescence detection based on previous descriptions (Schrumpf et al., 1991). Briefly, an HPLC system (Knauer Smartline Pump 1000 and Manager 5000 connected to Spark Triathlon autosampler) was used to mix 40 μL of sample solution with 360 μL of OPA reagent, of which 20 μL was loaded onto a reversed‐phase column with a guard column of the same material (MultoKrom 100–5 C18; CS‐Chromatographie Service GmbH, Langerwehe, Germany). At a flow rate of 1 mL/min, the samples were eluted over the course of 23 min using a gradient of the solvents Na‐acetate (50 mm, pH 7) and methanol as follows. Methanol was increased from initially 30% (1 min) to 70% over 6 min and then to 90% over 11 min. After a brief increase to 95% over 1 min, methanol was reduced to 70% over 2 min and then back to 30% over 2 min. After separation, the isoindole derivatives formed during OPA derivatization were detected with a fluorescence detector (Shimadzu RF‐10A XL, excitation: 330 nm, emission: 450 nm). Quantification was accomplished using commercial standards (Merck KGaA) and a representative chromatogram can be found in Figure S6A.
Compound identification was done via coupled UHPLC–MS/MS following a similar pre‐column OPA‐derivatization and separation protocol. For this, a Dionex UltiMate 3000 HPLC system (autosampler, pump and oven; Thermo Scientific, Bremen, Germany) with a Eurospher II 100–2 C18 column (Knauer GmbH, Berlin, Germany) coupled to a micrOTOF‐Q (Bruker Daltonics, Bremen, Germany) electrospray ionization and orthogonal time‐of‐flight mass spectrometer in the positive mode was employed. Representative chromatograms can be found in Figure S7.
SDS‐PAGE, Western blotting and immunostaining
Proteins were extracted and separated as described previously (Baier et al., 2018) using 10% SDS‐Polyacrylamide gels, followed by colloidal Coomassie staining (Dyballa and Metzger, 2009) and Western blotting onto a nitrocellulose membrane (AmershamTM; GE Healthcare, Uppsala, Sweden). After blocking with 2.5% (w/v) BSA and 2.5% (w/v) milk powder in TBS‐T buffer, immunodetection was carried out via incubation with horseradish peroxidase coupled Strep II tag (IBA Life Sciences, Göttingen, Germany) or HA‐tag antibodies (Thermo Scientific) in blocking buffer (1:10000), washing with TBS‐T and addition of Pierce™ ECL Western blotting substrates (Thermo Scientific), followed by detection in a Fusion Fx7 chemiluminescence imaging system (Vilber Lourmat).
Putrescine toxicity assay
A mixotrophic cultivation of strain UVM4 in a TAP medium was spiked with different amounts of putrescine hydrochloride (0.001 to 10 mm). Growth and cell morphology were observed for 4 days after which polyamines were extracted. Cell concentration was used to calculate the growth rate inhibition (GR) according to (Hafner et al., 2016). The GR value describes the ratio of growth rates in treated versus untreated conditions and is defined as 1 for no effect on growth and 0 for complete cytostasis.
Arginase assay
In vitro activity of recombinant arginases was quantified by colorimetric detection of the secondary enzymatic product urea. Total protein extraction was based on previous descriptions (Goldraij and Polacco, 1999). Briefly, about 2 × 107 cells were centrifuged at 1000 g for 3 min and the pellet was resuspended in 0.5 mL TBS buffer containing 4 mm Pefabloc® SC‐Protease‐Inhibitor (Carl Roth GmbH + Co. KG). About 0.3 mL of glass beads (0.1 mm; Carl Roth GmbH + Co. KG) was added and the sample was homogenized at 7500 rpm for 3 × 10 s with 15 s pauses using a cell homogenizer (Precellys Evolution; Bertin Technologies, Frankfurt am Main, Germany). The debris was centrifuged at 10000 g for 30 s and 300 μL of the supernatant was mixed with cofactor solution and after short incubation with the arginine substrate solution. The final reaction mixture (600 μL) contained about 0.4 mg/mL of protein (calculated from Hammel et al., 2018), 10 mm MnCl2, 160 mm L‐arginine at pH 9.7 and 200 μm of the urease inhibitor phenyl phosphorodiamidate (Liao and Raines, 1985). The sample was incubated for 30 min at 30 °C and the reaction was stopped by addition of 300 μL of 0.5 m H2SO4.
Colorimetric detection was based on the reaction of OPA, NED and urea at 37 °C as previously reported (Jung et al., 1975). About 200 μL of the final NED/OPA reagent (100 mg/L ortho‐phthalaldehyde, 215 mg/L N‐(1‐naphthyl)ethylenediamine (NED), 2.5 m sulphuric acid, 2.5 g/L boric acid and 0.03% Brij‐35 were mixed with 50 μL of sample solution in a 96‐well microtitre plate and incubated at room temperature in the dark for 30 min. Absorption at 505 nm was measured with a plate reader (Infinite M200 PRO, Tecan Group Ltd.).
Conflict of interest
The authors declare that they have no conflict of interest.
Author contributions
R.A.F.: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft. L.W., Data curation, Formal analysis, Investigation. A.E., Investigation. T.B., Conceptualization, Supervision, Writing – review and editing. O.K., Conceptualization, Funding acquisition, Project administration, Resources, Writing – review and editing.
Supporting information
Figure S1 Effect of extracellular putrescine on C. reinhardtii physiology.
Figure S2 Verification of disrupted target sites via cPCR.
Figure S3 Effects of ODC knockout and spermidine or putrescine supplementation.
Figure S4 Vector optimization for recombinant ODC overexpression.
Figure S5 Performance parameters of different ornithine decarboxylases.
Figure S6 Typical fluorescence chromatograms for amine compound quantification after RP‐HPLC and pre‐column OPA derivatization.
Figure S7 Typical UHPLC–MS/MS results for identification of 4‐aminobutanol.
Figure S8 Secondary product quantification during EcoSpeC and EcSpeF ODC overexpression.
Figure S9 Effect of substrate supplementation on putrescine accumulation.
Figure S10 Sequencing results of the AMX1 target region after successful Cas9‐RNP edit.
Figure S11 Effects of AMX gene knockouts on putrescine accumulation in mixotrophic conditions.
Figure S12 Phototrophic, high cell density cultivation of the reference strain N‐UVM4.
Figure S13 In vitro digest of different sgRNA target regions by Cas9‐RNPs.
Table S1 Sequence of all synthetic GOI coding sequences.
Table S2 Important primers that were used during this study.
Table S3 All tested sgRNA target sites.
Acknowledgements
This research was funded by the European Regional Development Fund (ERDF) and the Ministry of Economic Affairs, Innovation, Digitization and Energy of the State of North Rhine‐Westphalia by grant ‘Cluster Industrial Biotechnology (CLIB) Kompetenzzentrum Biotechnologie (CKB)’ (34.EFRE‐0300095/1703FI04). The authors would like to thank Prof. Dr. Ralph Bock for sharing strain UVM4 and want to acknowledge the contribution of Dr. Marcus Persicke and Alina Celine Tekin to the investigation. Open Access funding enabled and organized by Projekt DEAL.
References
- Atteia, A. , Adrait, A. , Brugire, S. , Tardif, M. , Van Lis, R. , Deusch, O. , Dagan, T. et al. (2009) A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the α‐proteobacterial mitochondrial ancestor. Mol. Biol. Evol. 26, 1533–1548. [DOI] [PubMed] [Google Scholar]
- Baier, T. , Jacobebbinghaus, N. , Einhaus, A. , Lauersen, K.J. and Kruse, O. (2020) Introns mediate post‐transcriptional enhancement of nuclear gene expression in the green microalga Chlamydomonas reinhardtii . PLoS Genet. 16, e1008944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier, T. , Wichmann, J. , Kruse, O. and Lauersen, K.J. (2018) Intron‐containing algal transgenes mediate efficient recombinant gene expression in the green microalga Chlamydomonas reinhardtii . Nucleic Acids Res. 46, 6909–6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold, P. , Schmitt, R. and Mages, W. (2002) An engineered Streptomyces hygroscopicus aph 7″ gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii . Protist 153, 401–412. [DOI] [PubMed] [Google Scholar]
- Bertoldi, M. , Carbone, V. and Borri Voltattorni, C. (1999) Ornithine and glutamate decarboxylases catalyse an oxidative deamination of their α‐methyl substrates. Biochem. J. 342, 509. [PMC free article] [PubMed] [Google Scholar]
- Blifernez‐Klassen, O. , Klassen, V. , Doebbe, A. , Kersting, K. , Grimm, P. , Wobbe, L. and Kruse, O. (2012) Cellulose degradation and assimilation by the unicellular phototrophic eukaryote Chlamydomonas reinhardtii . Nat. Commun. 3, 1214. [DOI] [PubMed] [Google Scholar]
- Calatrava, V. , Hom, E.F.Y. , Llamas, Á. , Fernández, E. and Galván, A. (2019) Nitrogen scavenging from amino acids and peptides in the model alga Chlamydomonas reinhardtii. The role of extracellular l‐amino oxidase. Algal Res. 38, 101395. [Google Scholar]
- Cao, M. , Fu, Y. , Guo, Y. and Pan, J. (2009) Chlamydomonas (Chlorophyceae) colony PCR. Protoplasma 235, 107–110. [DOI] [PubMed] [Google Scholar]
- Carmo‐Silva, A.E. , Keys, A.J. , Beale, M.H. , Ward, J.L. , Baker, J.M. , Hawkins, N.D. , Arrabaça, M.C. et al. (2009) Drought stress increases the production of 5‐hydroxynorvaline in two C4 grasses. Phytochemistry 70, 664–671. [DOI] [PubMed] [Google Scholar]
- Cerutti, H. , Johnson, A.M. , Gillham, N.W. and Boynton, J.E. (1997) A eubacterial gene conferring spectinomycin resistance on Chlamydomonas reinhardtii: integration into the nuclear genome and gene expression. Genetics 145, 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , McCaig, B.C. , Melotto, M. , He, S.Y. and Howe, G.A. (2004) Regulation of plant arginase by wounding, jasmonate, and the phytotoxin coronatine. J. Biol. Chem. 279, 45998–46007. [DOI] [PubMed] [Google Scholar]
- Chen, D. , Shao, Q. , Yin, L. , Younis, A. and Zheng, B. (2019) Polyamine function in plants: metabolism, regulation on development, and roles in abiotic stress responses. Front. Plant Sci. 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, E. , Arad, S. , Heimer, Y.M. and Mizrahi, Y. (1983) Polyamine biosynthetic enzymes in chlorella: Characterization of ornithine and arginine decarboxylase. Plant Cell Physiol. 24, 1003–1010. [Google Scholar]
- Crozet, P. , Navarro, F.J. , Willmund, F. , Mehrshahi, P. , Bakowski, K. , Lauersen, K.J. , Pérez‐Pérez, M.E. et al. (2018) Birth of a photosynthetic chassis: A MoClo toolkit enabling synthetic biology in the microalga Chlamydomonas reinhardtii . ACS Synth. Biol. 7, 2074–2086. [DOI] [PubMed] [Google Scholar]
- Dyballa, N. and Metzger, S. (2009) Fast and sensitive colloidal Coomassie G‐250 staining for proteins in polyacrylamide gels. J. Vis. Exp. 30. 10.3791/1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einhaus, A. , Baier, T. , Rosenstengel, M. , Freudenberg, R.A. and Kruse, O. (2021) Rational promoter engineering enables robust terpene production in microalgae. ACS Synth. Biol. 10, 847–856. [DOI] [PubMed] [Google Scholar]
- Einhaus, A. , Steube, J. , Freudenberg, R. , Barczyk, J. , Baier, T. and Kruse, O. (2022) Engineering a powerful green cell factory for robust photoautotrophic diterpenoid production. Metab. Eng. 73, 82–90. 10.1016/j.ymben.2022.06.002 [DOI] [PubMed] [Google Scholar]
- Fernandez, E. and Galvan, A. (2008) Nitrate assimilation in chlamydomonas. Eukaryot. Cell 7, 555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentino, G. , Zucaro, A. and Ulgiati, S. (2019) Towards an energy efficient chemistry. Switching from fossil to bio‐based products in a life cycle perspective. Energy 170, 720–729. [Google Scholar]
- Fischer, N. and Rochaix, J.‐D. (2001) The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii . Mol. Gen. Genomics. 265, 888–894. [DOI] [PubMed] [Google Scholar]
- Freudenberg, R.A. , Baier, T. , Einhaus, A. , Wobbe, L. and Kruse, O. (2021) High cell density cultivation enables efficient and sustainable recombinant polyamine production in the microalga Chlamydomonas reinhardtii . Bioresour. Technol. 323, 124542. [DOI] [PubMed] [Google Scholar]
- Freudenberg, R.A. , Wittemeier, L. , Einhaus, A. , Baier, T. and Kruse, O. (2022) The spermidine synthase gene SPD1: A novel auxotrophic marker for Chlamydomonas reinhardtii designed by enhanced CRISPR/Cas9 gene editing. Cell 11, 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldraij, A. and Polacco, J.C. (1999) Arginase is inoperative in developing soybean embryos. Plant Physiol. 119, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Ballester, D. , Sanz‐Luque, E. , Galván, A. , Fernández, E. and de Montaigu, A. (2018) Arginine is a component of the ammonium‐CYG56 signalling cascade that represses genes of the nitrogen assimilation pathway in Chlamydomonas reinhardtii . PLoS One 13, e0196167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman, D.S. and Levine, R.P. (1965) Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi . Proc. Natl. Acad. Sci. 54, 1665–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirard, B.M. and Snell, E.E. (1980) Purification and properties of ornithine decarboxylase from Lactobacillus sp. 30a. J. Biol. Chem. 255, 5960–5964. [PubMed] [Google Scholar]
- Hafner, M. , Niepel, M. , Chung, M. and Sorger, P.K. (2016) Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat. Methods 13, 521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel, A. , Zimmer, D. , Sommer, F. , Mühlhaus, T. and Schroda, M. (2018) Absolute quantification of major photosynthetic protein complexes in Chlamydomonas reinhardtii using quantification concatamers (QconCATs). Front. Plant Sci. 9, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E.H. , Stern, D. and Witman, G. (2009) The Chlamydomonas Sourcebook, 2nd edn. Oxford, UK: Academic Press. [Google Scholar]
- Hill, R.E. , White, R.L. and Smith, K.C. (1993) The identification of 5‐hydroxyl‐L‐norvaline in cultures of pyridoxine auxotrophs of Escherichia coli B. J. Nat. Prod. 56, 1246–1254. [DOI] [PubMed] [Google Scholar]
- Igarashi, K. and Kashiwagi, K. (2010) Modulation of cellular function by polyamines. Int. J. Biochem. Cell Biol. 42, 39–51. [DOI] [PubMed] [Google Scholar]
- Jaeger, D. , Baier, T. and Lauersen, K.J. (2019) Intronserter, an advanced online tool for design of intron containing transgenes. Algal Res. 42, 101588. [Google Scholar]
- Jancewicz, A.L. , Gibbs, N.M. and Masson, P.H. (2016) Cadaverine's functional role in plant development and environmental response. Front. Plant Sci. 7, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, D. , Biggs, H. , Erikson, J. and Ledyard, P.U. (1975) New colorimetric reaction for end point, continuous flow, and kinetic measurement of urea. Clin. Chem. 21, 1136–1140. [PubMed] [Google Scholar]
- Kindle, K.L. (1990) High‐frequency nuclear transformation of Chlamydomonas reinhardtii . Proc. Natl. Acad. Sci. 87, 1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk, D.L. and Kirk, M.M. (1978) Carrier‐mediated uptake of arginine and urea by Chlamydomonas reinhardtii . Plant Physiol. 61, 556–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat, J. , Hong‐Hermesdorf, A. , Casero, D. , Ent, P. , Castruita, M. , Pellegrini, M. , Merchant, S.S. et al. (2011) A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii . Plant J. 66, 770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labadorf, A. , Link, A. , Rogers, M.F. , Thomas, J. , Reddy, A.S. and Ben‐Hur, A. (2010) Genome‐wide analysis of alternative splicing in Chlamydomonas reinhardtii . BMC Genomics 11, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauersen, K.J. , Baier, T. , Wichmann, J. , Wördenweber, R. , Mussgnug, J.H. , Hübner, W. , Huser, T. et al. (2016) Efficient phototrophic production of a high‐value sesquiterpenoid from the eukaryotic microalga Chlamydomonas reinhardtii . Metab. Eng. 38, 331–343. [DOI] [PubMed] [Google Scholar]
- Lee, Y.S. and Cho, Y.D. (2001) Identification of essential active‐site residues in ornithine decarboxylase of Nicotiana glutinosa decarboxylating both L‐ornithine and L‐lysine. Biochem. J. 360, 657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. , Lowe‐Power, T. , Kurihara, S. , Gonzales, S. , Naidoo, J. , MacMillan, J.B. , Allen, C. et al. (2016) Functional identification of putrescine C‐ and N ‐Hydroxylases. ACS Chem. Biol. 11, 2782–2789. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Shen, Y.P. , Jiang, X.L. , Feng, L.S. and Liu, J.Z. (2018) Metabolic evolution and a comparative omics analysis of Corynebacterium glutamicum for putrescine production. J. Ind. Microbiol. Biotechnol. 45, 123–139. [DOI] [PubMed] [Google Scholar]
- Liao, C.F.‐H. and Raines, S.G. (1985) Inhibition of soil urease activity by amido derivatives of phosphoric and thiophosphoric acids. Plant Soil 85, 149–152. [Google Scholar]
- Lin, H.Y. and Lin, H.J. (2019) Polyamines in microalgae: something borrowed, something new. Mar. Drugs 17. 10.3390/md17010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Paz, C. , Liu, D. , Geng, S. and Umen, J.G. (2017) Identification of Chlamydomonas reinhardtii endogenous genic flanking sequences for improved transgene expression. Plant J. 92, 1232–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppes, R. (1970) Growth inhibition by NH4+ ions in arginine‐requiring mutants of Chlamydomonas reinhardi . MGG Mol. Gen. Genet. 109, 233–240. [DOI] [PubMed] [Google Scholar]
- McFadden, G.I. and Melkonian, M. (1986) Use of Hepes buffer for microalgal culture media and fixation for electron microscopy. Phycologia 25, 551–557. [Google Scholar]
- Merchant, S.S. , Prochnik, S.E. , Vallon, O. , Harris, E.H. , Karpowicz, S.J. , Witman, G.B. , Terry, A. et al. (2007) The chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, A.J. (2016a) Polyamines in eukaryotes, bacteria, and archaea. J. Biol. Chem. 291, 14896–14903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, A.J. (2016b) Biosynthesis of polyamines and polyamine‐containing molecules. Biochem. J. 473, 2315–2329. [DOI] [PubMed] [Google Scholar]
- Miller‐Fleming, L. , Olin‐Sandoval, V. , Campbell, K. and Ralser, M. (2015) Remaining mysteries of molecular biology: the role of polyamines in the cell. J. Mol. Biol. 427, 3389–3406. [DOI] [PubMed] [Google Scholar]
- Munz, J. , Xiong, Y. , Kim, J.Y.H. , Sung, Y.J. , Seo, S. , Hong, R.H. , Kariyawasam, T. et al. (2020) Arginine‐fed cultures generates triacylglycerol by triggering nitrogen starvation responses during robust growth in Chlamydomonas. Algal Res. 46, 101782. [Google Scholar]
- Neupert, J. , Karcher, D. and Bock, R. (2009) Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 57, 1140–1150. [DOI] [PubMed] [Google Scholar]
- Pandey, M. , Stormo, G.D. and Dutcher, S.K. (2020) Alternative splicing during the Chlamydomonas reinhardtii cell cycle. G3 Genes¦Genomes¦Genetics 10, 3797–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.J. , Wang, H. , Gargouri, M. , Deshpande, R.R. , Skepper, J.N. , Holguin, F.O. , Juergens, M.T. et al. (2015) The response of Chlamydomonas reinhardtii to nitrogen deprivation: a systems biology analysis. Plant J. 81, 611–624. [DOI] [PubMed] [Google Scholar]
- Poidevin, L. , Unal, D. , Belda‐Palazón, B. and Ferrando, A. (2019) Polyamines as quality control metabolites operating at the post‐transcriptional level. Plan. Theory 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontrelli, S. , Fricke, R.C.B. , Teoh, S.T. , Laviña, W.A. , Putri, S.P. , Fitz‐Gibbon, S. , Chung, M. et al. (2018) Metabolic repair through emergence of new pathways in Escherichia coli . Nat. Chem. Biol. 14, 1005–1009. [DOI] [PubMed] [Google Scholar]
- Prabowo, C.P.S. , Shin, J.H. , Cho, J.S. , Chae, T.U. and Lee, S.Y. (2020) Microbial production of 4‐amino‐1‐butanol, a four‐carbon amino alcohol. Biotechnol. Bioeng. 117, 2771–2780. [DOI] [PubMed] [Google Scholar]
- Rangan, P. , Subramani, R. , Kumar, R. , Singh, A.K. and Singh, R. (2014) Recent advances in polyamine metabolism and abiotic stress tolerance. Biomed. Res. Int. 2014, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roerdink, E. and Warnier, J.M.M. (1985) Preparation and properties of high molar mass nylon‐4,6: a new development in nylon polymers. Polymer 26, 1582–1588. [Google Scholar]
- Sager, R. and Granick, S. (1953) Nutritional studies with Chlamydomonas reinhardi . Ann. N. Y. Acad. Sci. 56, 831–838. [DOI] [PubMed] [Google Scholar]
- Sakai, K. , Miyasako, Y. , Nagatomo, H. , Watanabe, H. , Wakayama, M. and Moriguchi, M. (1997) L‐ornithine decarboxylase from Hafnia alvei has a novel L‐ornithine oxidase activity. J. Biochem. 122, 961–968. [DOI] [PubMed] [Google Scholar]
- Sasikumar, K. , Hannibal, S. , Wendisch, V.F. and Nampoothiri, K.M. (2021) Production of biopolyamide precursors 5‐amino valeric acid and putrescine from rice straw hydrolysate by engineered Corynebacterium glutamicum . Front. Bioeng. Biotechnol. 9, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmollinger, S. , Mühlhaus, T. , Boyle, N.R. , Blaby, I.K. , Casero, D. , Mettler, T. , Moseley, J.L. et al. (2014) Nitrogen‐sparing mechanisms in chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 26, 1410–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, J. and Wendisch, V.F. (2010) Putrescine production by engineered Corynebacterium glutamicum . Appl. Microbiol. Biotechnol. 88, 859–868. [DOI] [PubMed] [Google Scholar]
- Schneider, J. and Wendisch, V.F. (2011) Biotechnological production of polyamines by Bacteria: recent achievements and future perspectives. Appl. Microbiol. Biotechnol. 91, 17–30. [DOI] [PubMed] [Google Scholar]
- Schroda, M. , Blocker, D. and Beck, C.F. (2000) The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 21, 121–131. [DOI] [PubMed] [Google Scholar]
- Schrumpf, B. , Schwarzer, A. , Kalinowski, J. , Puhler, A. , Eggeling, L. and Sahm, H. (1991) A functionally split pathway for lysine synthesis in Corynebacterium glutamicum . J. Bacteriol. 173, 4510–4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selim, K.A. , Ermilova, E. and Forchhammer, K. (2020a) From cyanobacteria to Archaeplastida: new evolutionary insights into PII signalling in the plant kingdom. New Phytol. 227, 722–731. [DOI] [PubMed] [Google Scholar]
- Selim, K.A. , Lapina, T. , Forchhammer, K. and Ermilova, E. (2020b) Interaction of N‐acetyl‐l‐glutamate kinase with the PII signal transducer in the non‐photosynthetic alga Polytomella parva: co‐evolution towards a hetero‐oligomeric enzyme. FEBS J. 287, 465–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, S. , Guo, S.‐R. and Yu, L.‐Y. (2012) A review: polyamines and photosynthesis In advances in photosynthesis – fundamental aspects, pp. 441–445. INTECH. 10.5772/26875 [DOI] [Google Scholar]
- Siddappa, S. and Marathe, G.K. (2020) What we know about plant arginases? Plant Physiol Biochemistry 156, 600–610. [DOI] [PubMed] [Google Scholar]
- Sizova, I. , Fuhrmann, M. and Hegemann, P. (2001) A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii . Gene 277, 221–229. [DOI] [PubMed] [Google Scholar]
- Slocum, R.D. (2005) Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Physiol. Biochem. 43, 729–745. [DOI] [PubMed] [Google Scholar]
- Stevens, D.R. , Rochaix, J.D. and Purton, S. (1996) The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet. 251, 23–30. [DOI] [PubMed] [Google Scholar]
- Stiti, N. , Missihoun, T.D. , Kotchoni, S.O. , Kirch, H.‐H. and Bartels, D. (2011) Aldehyde dehydrogenases in Arabidopsis thaliana: biochemical requirements, metabolic pathways, and functional analysis. Front. Plant Sci. 2, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussenbach, J.S. and Strijkert, P.J. (1969) Arginine metabolism in Chlamydomonas reinhardi. On the regulation of the arginine biosynthesis. Eur. J. Biochem. 8, 403–407. [DOI] [PubMed] [Google Scholar]
- Takatsuka, Y. , Tomita, T. and Kamio, Y. (1999) Identification of the amino acid residues conferring substrate specificity upon Selenomonas ruminantium lysine decarboxylase. Biosci. Biotechnol. Biochem. 63, 1843–1846. [DOI] [PubMed] [Google Scholar]
- Tassoni, A. , Awad, N. and Griffiths, G. (2018) Effect of ornithine decarboxylase and norspermidine in modulating cell division in the green alga Chlamydomonas reinhardtii . Plant Physiol. Biochem. 123, 125–131. [DOI] [PubMed] [Google Scholar]
- Theiss, C. , Bohley, P. , Bisswanger, H. and Voigt, J. (2004) Uptake of polyamines by the unicellular green alga Chlamydomonas reinhardtii and their effect on ornithine decarboxylase activity. J. Plant Physiol. 161, 3–14. [DOI] [PubMed] [Google Scholar]
- Theiss, C. , Bohley, P. and Voigt, J. (2002) Regulation by polyamines of ornithine decarboxylase activity and cell division in the unicellular green alga Chlamydomonas reinhardtii . Plant Physiol. 128, 1470–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinyard, W.A. , Fleming, A.M. , Ma, J. and Burrows, C.J. (2018) Characterization of G‐Quadruplexes in Chlamydomonas reinhardtii and the effects of polyamine and magnesium cations on structure and stability. Biochemistry 57, 6551–6561. [DOI] [PubMed] [Google Scholar]
- Vogel, R.H. and Kopac, M.J. (1959) Glutamic γ‐semialdehyde in arginine and proline synthesis of neurospora: a mutant‐tracer analysis. Biochim. Biophys. Acta 36, 505–510. [Google Scholar]
- Voigt, J. , Deinert, B. and Bohley, P. (2000) Subcellular localization and light‐dark control of ornithine decarboxylase in the unicellular green alga Chlamydomonas reinhardtii . Physiol. Plant. 108, 353–360. [Google Scholar]
- Wichmann, J. , Baier, T. , Wentnagel, E. , Lauersen, K.J. and Kruse, O. (2018) Tailored carbon partitioning for phototrophic production of (E)‐α‐bisabolene from the green microalga Chlamydomonas reinhardtii . Metab. Eng. 45, 211–222. [DOI] [PubMed] [Google Scholar]
- Yang, X. , Peng, J. and Pan, J. (2019) Nourseothricin N‐acetyl transferase (NAT), a new selectable marker for nuclear gene expression in Chlamydomonas. Plant Methods 15, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J.J. , Park, K.B. , Kim, S.G. and Oh, S.H. (2013) Expression, purification, and biochemical properties of arginase from Bacillus subtilis 168. J. Microbiol. 51, 222–228. [DOI] [PubMed] [Google Scholar]
- Zalutskaya, Z. , Derkach, V. , Puzanskiy, R. and Ermilova, E. (2020) Impact of nitric oxide on proline and putrescine biosynthesis in chlamydomonas via transcriptional regulation. Biol. Plant. 64, 642–648. [Google Scholar]
- Zalutskaya, Z. , Kochemasova, L. and Ermilova, E. (2018) Dual positive and negative control of Chlamydomonas PII signal transduction protein expression by nitrate/nitrite and NO via the components of nitric oxide cycle. BMC Plant Biol. 18, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, T. , Li, S. , Wang, J. , Zhou, Q. , Yang, C. , Bai, F. , Lan, X. et al. (2020) Engineering tropane alkaloid production based on metabolic characterization of ornithine decarboxylase in atropa belladonna. ACS Synth. Biol. 9, 437–448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Effect of extracellular putrescine on C. reinhardtii physiology.
Figure S2 Verification of disrupted target sites via cPCR.
Figure S3 Effects of ODC knockout and spermidine or putrescine supplementation.
Figure S4 Vector optimization for recombinant ODC overexpression.
Figure S5 Performance parameters of different ornithine decarboxylases.
Figure S6 Typical fluorescence chromatograms for amine compound quantification after RP‐HPLC and pre‐column OPA derivatization.
Figure S7 Typical UHPLC–MS/MS results for identification of 4‐aminobutanol.
Figure S8 Secondary product quantification during EcoSpeC and EcSpeF ODC overexpression.
Figure S9 Effect of substrate supplementation on putrescine accumulation.
Figure S10 Sequencing results of the AMX1 target region after successful Cas9‐RNP edit.
Figure S11 Effects of AMX gene knockouts on putrescine accumulation in mixotrophic conditions.
Figure S12 Phototrophic, high cell density cultivation of the reference strain N‐UVM4.
Figure S13 In vitro digest of different sgRNA target regions by Cas9‐RNPs.
Table S1 Sequence of all synthetic GOI coding sequences.
Table S2 Important primers that were used during this study.
Table S3 All tested sgRNA target sites.


