Figure 2.
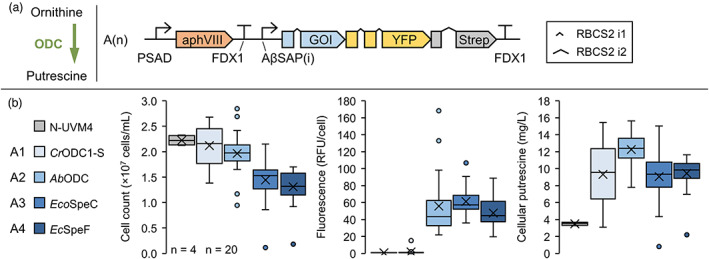
Establishing recombinant putrescine production in Chlamydomonas reinhardtii by recombinant expression of ornithine decarboxylases. (a) Decarboxylation reaction from ornithine to putrescine (left) and transgene design used in this experiment (right). (b) Strains with highest reporter fluorescence levels were cultivated mixotrophically for 4 days and compared to the parental strain N‐UVM4. The box‐and‐whisker plots depict the final cell concentration on day 4, adjusted YFP fluorescence level per cell and the volumetric cellular putrescine yield (from left to right) depending on the gene of interest (GOI). The number of biological replicates is indicated at the bottom left (n = 4 for N‐UVM4, n = 20 for A1‐A4). The number of introns in the GOI from A1 to A4 equals 4, 3, 5 and 5. Genetic symbols are based on the SBOL 3.0 suggestions or have been published previously (Crozet et al., 2018). The glyphs for promoter and terminator do include 5′ and 3′ UTRs, respectively. RFU, relative fluorescence units; PSAD Photosystem I reaction centre subunit II; FDX1, Ferredoxin; RBCS2 i1/i2, Ribulose bisphosphate carboxylase small subunit intron 1/2; AβSAP(i), HSP70A/βTUB2 Synthetic Algae Promoter; aphVIII, Aminoglycoside 3′‐phosphotransferase type VIII. [Colour figure can be viewed at wileyonlinelibrary.com]
