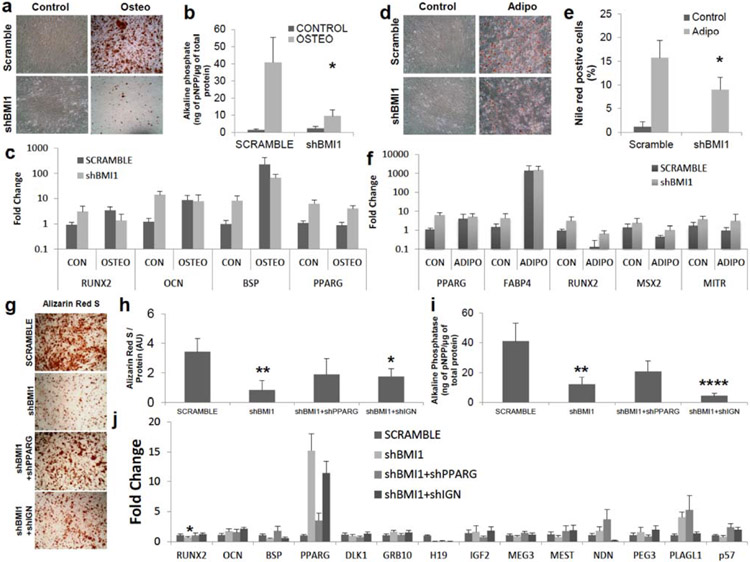
Fig. (3). Derepression of developmental regulator network via BMI1 knockdown decreases osteogenic and adipogenic differentiation.
(a-c) Osteogenesis in shBMI1 MSCs was tested. Control; no osteogenic induction, Osteo; osteogenic induction (a) Images from Alizarin Red S staining for osteoblasts. MSCs were cultured with osteogenic media for 14 days. (b) Alkaline phosphate activity (normalized by whole protein concentration) in shBMI1 was reduced (P=0.01), n=6. (c) mRNA levels of RUNX2, OCN and BSP in osteogenic differentiation, normalized as in Fig 1c, were reduced upon shBMI1 and derepression of RUNX2, PPARγ and OCN was observed without an osteogenic induction. n=3. PPARγ mRNA levels, well-known osteogenic inhibitor, were upregulated with and without osteogenic induction. (d-f) Adipogenesis in shBMI1 was evaluated 14 days after the treatment of adipogenic cocktails. Adipo; adipogenic induction (d) Images from Oil Red O staining for adipogenesis. (e) Nile Red+ cells by flow cytometry showed slight reductions in shBMI1 (P=0.049). n=4. (f) mRNA expressions of PPARγ and FABP4, normalized as in Fig 1b, shows that derepression of these transcripts were observed without adipogenic inductions. Gene expression of RUNX2, MSX2 and MITR , well-known osteogenic-related factors for blocking adipogenesis were evaluated. n=3. (g-j) PPARγ and IGN were necessary to rescue osteogenesis in shBMI1 MSCs (g) Images from Alizarin Red S staining for osteoblasts. Vector transduced MSCs were cultured with osteogenic media for 14 days. (h) Alizarin Red S was measured by acetic acid precipitation, normalized by whole protein concentration. P-value was calculated based on SCRAMBLE. shBMI1 (P=0.004), shBMI1+shIGN (P=0.04). AU; arbitrary unit. n=4. (i) Alkaline phosphate activity, normalized by whole protein concentration. p-value was calculated based on SCRAMBLE. shBMI1 (P=0.007), shBMI1+shIGN (P=0.0008). n=4. (j) mRNA levels of RUNX2, OCN and BSP in osteogenic differentiation, normalized to scramble osteogenic induction as 1, were shown. PPARγ mRNA levels, well-known osteogenic inhibitor, were upregulated upon shBMI1 and slight reduction in shIGN samples against shBMI1 alone. IGN transcripts were evaluated. n=4. *: P<0.05, **: P<0.01, ***: P<0.001, ****: P<0.0005 Data shown as mean ± SEM.