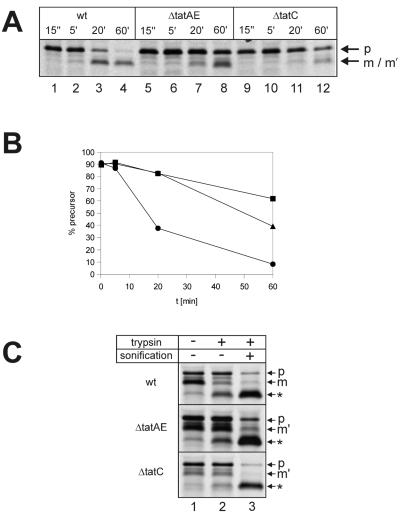
FIG. 5.
Processing of TorA-GFOR in E. coli tat mutants and localization of GFOR gene products by trypsin treatment of spheroplasts. (A) Pulse-chase experiments with E. coli strains MC4100, MC4100 ΔtatC, and MC4100 ΔtatAE, containing plasmid pTW42, encoding the TorA-GFOR fusion protein, were performed as described in the legend to Fig. 2. (B) The bands of the gel in panel A were quantified using a PhosphorImager and the FragmeNT Analysis (version 1.1) software (Molecular Dynamics). The percentages of precursor present at the indicated chase times were calculated [p/(p + m) × 100, where p is the amount of precursor and m is the amount of mature form]. Circles, MC4100 wild type; triangles, MC4100 tatAE mutant; squares, MC4100 tatC mutant. (C) To examine the subcellular localization of the different GFOR forms in E. coli MC4100, MC4100 ΔtatAE, and MC4100 ΔtatC, labeled cells were converted to spheroplasts and submitted to tryptic digestion as described in the legend to Fig. 3, with the exception that, due to the slower processing of TorA-GFOR in E. coli MC4100, cells were converted to spheroplasts after a 60-min chase. wt, wild type; p, precursor; m, mature GFOR; m′, mature-sized cytosolic GFOR fragment; ∗, tryptic fragment of GFOR.