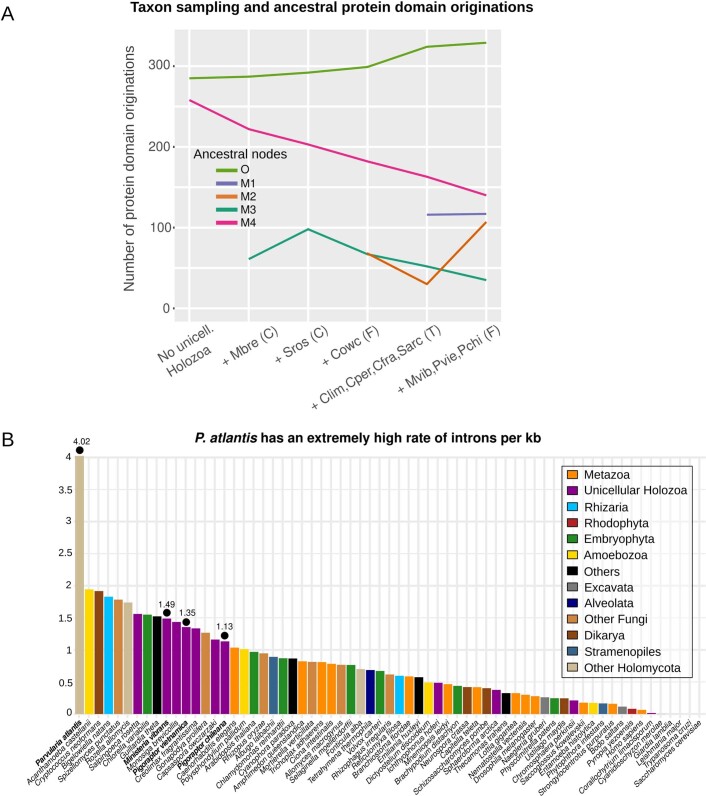
Extended Data Fig. 1. The importance of taxon sampling in ancestral gene content reconstructions and intron density across eukaryotes.
(A) Influence of taxon sampling in the ancestral reconstruction of protein domains innovations (Pfam domains). Note that with the addition of taxon sampling from unicellular relatives of animals (Choanoflagellatea -C-, Filasterea -F-, Teretosporea -T-), the number of pre-metazoan protein domain originations increase at the expense of originations that were originally detected at M4 in the 'No unicell. Holozoa' condition. The origin of every protein domain was inferred at the last common ancestor of all the species in which the domain is represented. This analysis was carried out with the taxon sampling euk_db, first excluding all representatives from C, F and T groups ('No unicell. Holozoa'), and then progressively adding data from these groups in a chronological order corresponding to when the genomic data from the representatives of these groups became publicly available. Ancestral node abbreviations: M4 = last common ancestor (LCA) of Metazoa. M3 = LCA of Choanoflagellatea and M4. M2 = LCA of Filasterea and M3. M1 = LCA of Teretosporea and M2. O = LCA of Opisthokonta. (See Fig. 1d for an illustration of the phylogenetic context of these ancestral nodes). (B) Distribution of introns per kb in an eukaryotic dataset including the four genomes sequenced for this manuscript as well as the metrics included in the Fig. 1—source data 1 of ref. 18.