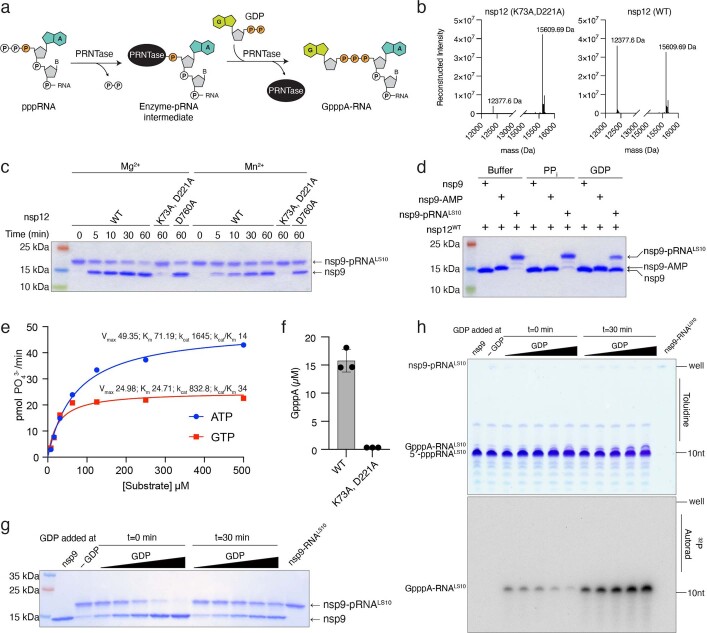
Extended Data Fig. 5. Characterization of nsp12 NiRAN GDP-PRNTase activity.
a. Schematic representation depicting the mechanism of core cap formation by vesicular stomatitis virus (VSV) polyribonucleotidyltransferase (PRNTase). b. Intact mass LC/MS spectra of nsp9–pRNALS10 after incubation with GDP and WT nsp12 (right) or the NiRAN mutant (left). The theoretical mass of nsp9 is 12378.2 Da and the theoretical mass of nsp9–pRNALS10 is 15611.5 Da. c. Time-dependent deRNAylation of nsp9–pRNALS10 by WT nsp12, the NiRAN mutant (K73A, D221A), or the polymerase mutant (D760A) in the presence of GDP and either Mg2+ or Mn2+. Reaction products were analysed as in Fig. 2b. d. NiRAN-dependent deAMPylation or deRNAylation of nsp9 in the presence of PPi or GDP. Reaction products were analysed as in Fig. 2b. e. Concentration dependence of ATP (blue) or GTP (red) on the rate of phosphate release catalysed by nsp13. Vmax (pmol PO43−/min), Km (μM), kcat (min−1) and kcat/Km (min−1/μM) values are indicated. f. HPLC/MS quantification of GpppA formed during the NiRAN-catalysed deRNAylation of nsp9–pRNALS10. Reaction products were digested with nuclease P1 prior to HPLC analysis. Reactions were performed in triplicate and error bars represent the standard deviation. g, h. NiRAN-catalysed capping reactions depicting the inhibitory effect of GDP on RNAylation. nsp9 was incubated with excess 5′-pppRNALS10 in presence of nsp12 with no GDP (-GDP), or increasing concentrations (6.25-100 µM) of [α-32P]GDP added either at time zero (t = 0 min), or after the RNAylation reaction was allowed to proceed for 30 min (t=30 min). Reaction products were analysed by SDS-PAGE (g), and urea–PAGE (h).