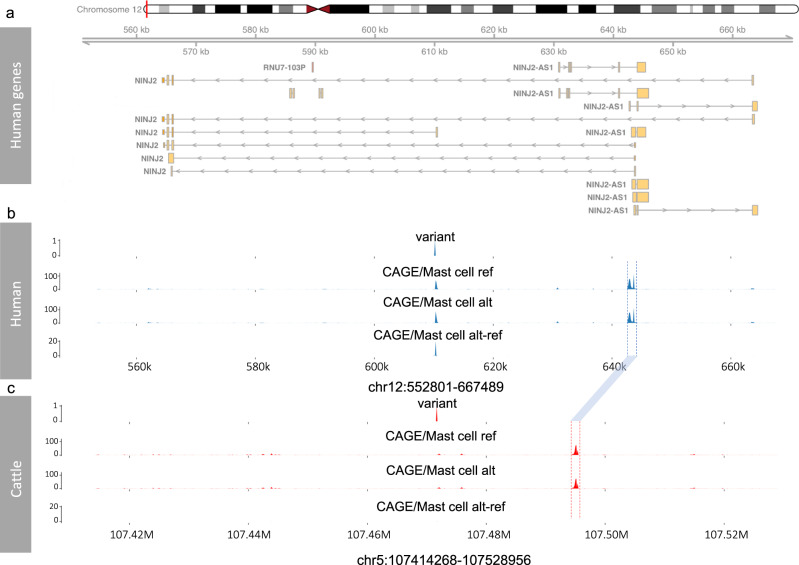
Fig. 8. Enformer predictions for human and cattle.
a The human gene neighborhood of variant rs10849334. Different isoforms of the genes are included in the plot. b Enformer predicted Cap Analysis Gene Expression (CAGE) tracks of variant rs10849334 in mast cells (the track showing the largest human alt-ref difference). The first three tracks show i) the position of the target variant in the human genome, and the predicted CAGE levels for the ii) reference and iii) alternative alleles from Enformer, i.e., the predicted CAGE tracks obtained by taking the human DNA sequences containing reference and alternative alleles at the variant position as the inputs of the Enformer model. The final track shows the predicted difference between the CAGE levels from the reference and alternate sequences (alt-ref). As can be seen the only difference is observed specifically at the start position of the shorter NINJ2 isoform in the centre of the plot. c Corresponding predicted CAGE tracks derived from the cattle sequences around the orthologous variant of rs10849334. The orthologous peaks at the TSSs of the longer NINJ2 transcripts in cattle and human are indicated by the blue linking bar. However, no CAGE peak is predicted at the TSS of the shorter isoform of NINJ2, unlike with the human sequences. The data underlying this figure can be found in Supplementary Data 8.