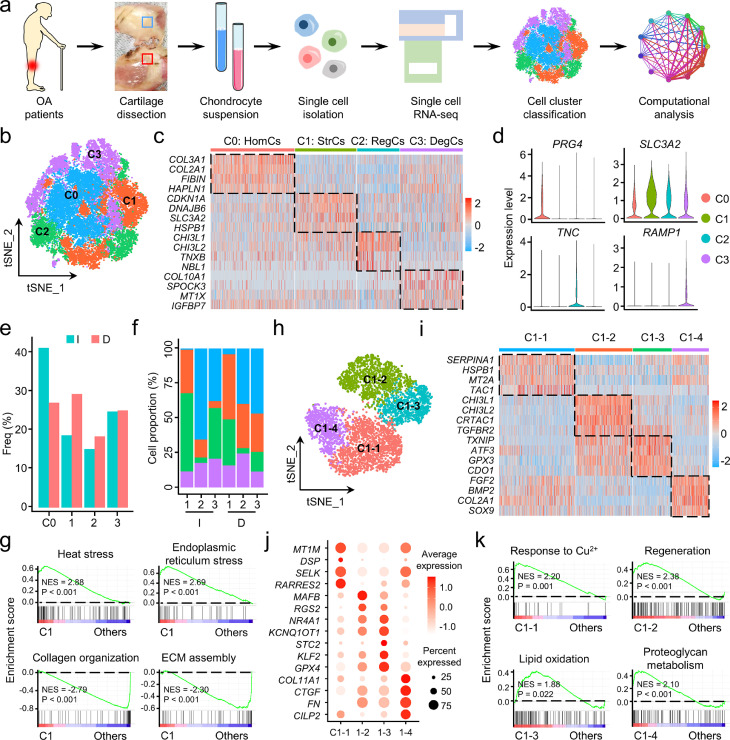
Figure 2.
Single-cell transcriptomic analysis of intact and damaged human osteoarthritis (OA) cartilages. (a) Schematic workflow of the experimental strategy. (b) The t-distributed stochastic neighbor embedding (t-SNE) plot of the four identified main chondrocyte clusters in OA cartilage. (c) Heatmap revealing the scaled expression of preferentially and differentially expressed genes for each chondrocyte cluster. (d) Violin plots demonstrating the normalized gene expression levels of representative marker genes among the four main clusters. (e) The frequency of each chondrocyte cluster in the intact (I) and damaged (D) human OA cartilages. Freq, frequency. (f) Relative proportion of each chondrocyte cluster across three pairs of intact (I) and damaged (D) cartilages as indicated. (g) Gene set enrichment analysis (GSEA) revealing the enrichment of Gene Ontology (GO) terms in C1 and the other three clusters. NES, normalized enrichment score; ECM, extracellular matrix. (h) Visualization of t-SNE colored subclusters within C1. (i) Heatmap showing the highly expressed genes in the four subclusters within C1. (j) Dot plot showing the mean expression of preferentially expressed genes among the four subclusters. (k) GSEA showing the representative function among each subcluster. NES, normalized enrichment score.