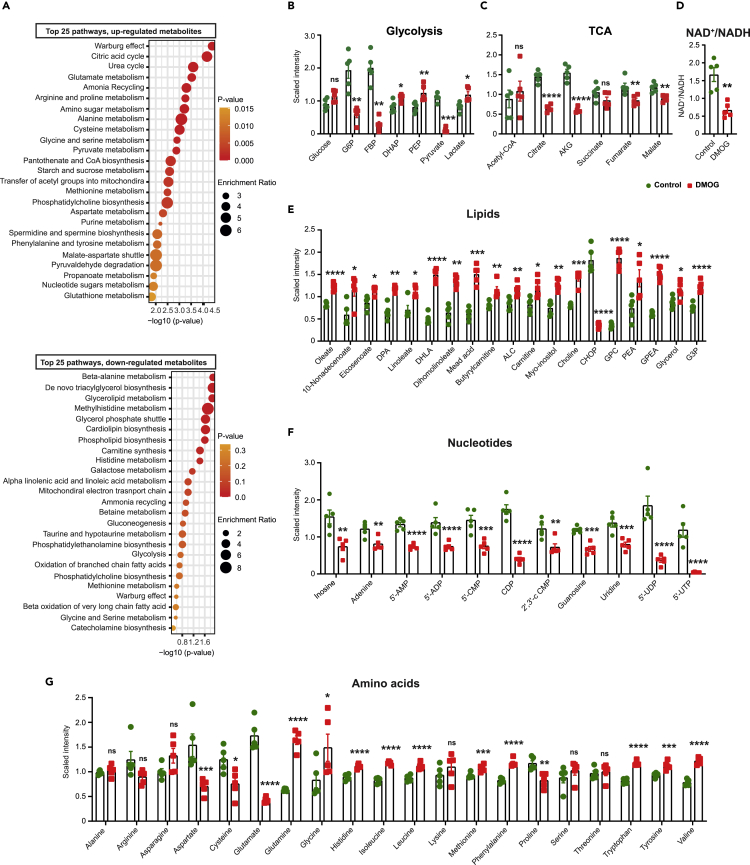
Figure 3.
DMOG alters the endothelial cell metabolome
(A) Shown are the top 25 downregulated (upper graph) and upregulated (lower graph) metabolic pathways detected by metabolites set enrichment analysis in DMOG-treated cells compared to control. Scaled intensity values indicating relative levels of metabolites related to glycolysis (B) and TCA cycle (C). (D) NAD+/NADH ratio in cells treated with vehicle or DMOG. Scaled intensity values indicating relative levels of lipid metabolites (E), nucleotides (F), and amino acids (G). n = 5 independent samples per condition. All statistical data are represented as mean ± SEM and statistics were determined by a Welch’s two sample t-test. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001; ns, not significant. G6P, glucose-6-phosphate; FBP, fructose 1,6 bisphosphate; DHAP, dihydroxyacetone phosphate; PEP, phosphoenolpyruvate; AKG, alpha-ketoglutarate; DPA, docosapentaenoate; DHLA, dihomolinolenate; ALC, acetylcarnitine; CHOP, choline phosphate; GPC, glycerophosphorylcholine; PEA, phosphoethanolamine; GPEA, glycerylphosphorylethanolamine; G3P, glycerol 3-phosphate; 5′-AMP, adenosine-5′-monophosphate; 5′-ADP, adenosine-5′-diphopshate; 5′-CMP, cytidine 5′-monophosphate; CDP, cytidine diphosphate; 2′,3′-cCMP, cytidine 2′,3′-cyclic monophosphate; 5′-UDP, uridine-5-diphosphate; UTP, uridine 5′-triphosphate. See also Figure S4 and Table S1.