SUMMARY
Spermidine is essential for cellular growth and acts as a prerequisite of hypusination, a post-translational modification of eukaryotic initiation factor 5A (eIF5A), allowing the translation of polyproline-containing proteins. Here, we show that oncogenic Kaposi’s sarcoma-associated herpesvirus (KSHV) increases spermidine synthesis and eIF5A hypusination to enhance expression of polyproline-containing latency-associated nuclear antigen (LANA) for viral episomal maintenance. KSHV upregulates intracellular spermidine levels by dysregulating polyamine metabolic pathways in three-dimensional (3D) culture and 2D de novo infection conditions. Increased intracellular spermidine leads to increased eIF5A hypusination, ultimately enhancing LANA expression. In contrast, inhibition of spermidine synthesis or eIF5A hypusination alleviates LANA expression, decreasing viral episomal maintenance and KSHV-infected cell proliferation in vitro and in vivo, which is reversed by spermidine supplement. This demonstrates that KSHV hijacks spermidine synthesis and eIF5A hypusination pathways to enhance LANA expression for viral episomal maintenance, suggesting polyamine metabolism and eIF5A hypusination as therapeutic targets for KSHV-induced tumorigenesis.
In brief
Human oncogenic viruses reprogram the host metabolic pathway for viral tumorigenesis. Choi et al. demonstrate that oncogenic Kaposi’s sarcoma-associated herpesvirus promotes eIF5A hypusination by enhancing spermidine synthesis, which is critical for viral episome maintenance. This suggests that the spermidine-eIF5A hypusination axis can be a potential target for anti-KSHV drug development.
Graphical Abstract
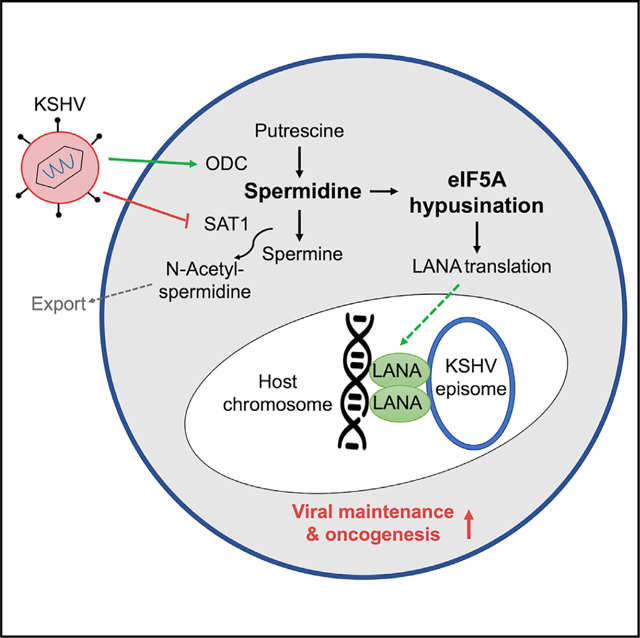
INTRODUCTION
Cellular metabolism is a highly regulated system that safeguards the homeostasis of our body. Aberrant metabolism can thus be associated with pathological consequences including cancer. Polyamines, which comprise putrescine, spermidine, and spermine, are highly charged polycationic alkylamines that are abundantly found in all living cells (Agostinelli et al., 2010). These cationic metabolites tend to interact with DNA and contribute to DNA stabilization, modulation of chromatin structure, and gene transcription (Gerner and Meyskens, 2004; Pegg, 2009). Intact regulation of the polyamine metabolic pathway is imperative in keeping the cells functional, as it plays a critical role in fundamental biological processes, including cellular proliferation and protein and nucleic acid syntheses (Puleston et al., 2021; Schroeder et al., 2021). Thus, aberrant upregulation of this pathway has been shown in rapidly growing cells, such as cancer cells (Casero and Marton, 2007; Khan et al., 2021). The level of cellular polyamines is controlled by their synthesis, degradation, and efflux system. As the first rate-limiting enzyme in the polyamine synthesis pathway, ornithine decarboxylase (ODC) converts ornithine to putrescine (Pegg, 2006). Subsequently, putrescine is converted to spermidine and spermine by the addition of an aminopropyl moiety derived from methionine. The polyamine catabolic process is governed by enzymes called spermidine/spermine N1-acetyltransferase (SAT1) that acetylates spermidine and spermine, respectively, to produce N-acetylspermidine and N-acetylspermine (Pegg, 2008). These acetylated polyamines can be exported out of the cells, thus preventing excessive polyamine accumulation and maintaining polyamine homeostasis. This polyamine homeostasis is disrupted in cancer cells through c-Myc, mutant adenomatous polyposiscoli (APC), or KRAS oncogenes that either increase the polyamine synthesis or suppress the polyamine catabolism to promote tumorigenesis (Bello-Fernandez et al., 1993; Erdman et al., 1999; Ignatenko et al., 2004).
Among the three polyamines, spermidine serves as a substrate for hypusination, a post-translational modification process catalyzed by enzymes called deoxyhypusine synthase (DHPS) and deoxyhypusine hydroxylase (DOHH). To date, eukaryotic translation initiation factor 5A (eIF5A) is the only known protein that undergoes hypusination modification (Park, 2006). In addition to its role in promoting elongation and termination, eIF5A is a critical factor in global translation, as it is required in relieving stalled ribosomes that are necessary for continuous translation of mRNAs (Pelechano and Alepuz, 2017; Schuller et al., 2017). Previous genome-wide analyses of ribosome stalling events in eIF5A-deficient cells identified multiple stalling-prone amino acid sequences, including proline, glycine, and aspartic acid (Pelechano and Alepuz, 2017; Ude et al., 2013). In particular, hypusination of eIF5A is essential for efficient translation of proteins containing polyproline sequences, as hypusine moiety of eIF5A interacts with proline-tRNA and stabilizes an optimal geometric structure for peptide bond formation (Gutierrez et al., 2013; Huter et al., 2017; Schmidt et al., 2016). Hypusinated eIF5A is also implicated in tumorigenesis by increasing the translation of c-Myc and KRAS oncoproteins (Fujimura et al., 2018). Accordingly, blockade of eIF5A hypusination has been shown to reduce tumor cell growth in vitro and decrease polyp size in a mouse colon cancer model (Coni et al., 2020; Nakanishi and Cleveland, 2016).
Kaposi’s sarcoma-associated herpesvirus (KSHV) is the etiologic agent of Kaposi’s sarcoma (KS) and primary effusion lymphoma (PEL), which are highly prevalent in AIDS patients (Chang et al., 1994). Aggressive endemic KS, primarily seen in Africa, accounts for nearly half of the reported cancers and is among the leading cause of cancer death within the region. KSHV infection is predominantly latent in tumor cells, with only a limited number of cells undergoing lytic replication. During latency, KSHV only expresses a small subset of viral proteins, among which include the major latent protein, latency-associated nuclear antigen (LANA) (Decker et al., 1996; Parravicini et al., 2000). LANA functions as a molecular tether that bridges the viral episome to the host mitotic chromosomes to ensure segregation of KSHV episomes to the progeny nuclei (Ballestas et al., 1999). Thus, LANA is essential for transformation, latency maintenance, episomal DNA replication, and segregation in KSHV-infected tumor cells (Ye et al., 2004). Molecularly, the N-terminal proline-rich region of LANA attaches to the host chromosomes while its C-terminal region binds the viral terminal repeat sequences and the central acid-rich repeat sequence to ensure efficient episome maintenance (De Leon Vazquez and Kaye, 2011; Komatsu et al., 2004; Srinivasan et al., 2004). Overall, these prominent features of LANA as a crucial mediator of virus persistence make it an attractive therapeutic target against KSHV-associated cancers.
The metabolic properties of KSHV-infected cells closely resemble the metabolic hallmarks of cancer cells (Levy and Bartosch, 2016). To date, most metabolic studies have relied on two-dimensional cell-culture systems, which have limitations in faithfully recapitulating the characteristic features of KSHV-associated tumors. Primarily, 2D cell-culture systems fail to mimic essential tumor microenvironments, such as cell-to-cell and cell-extracellular matrix (ECM) interactions that affect tumorigenesis (Holle et al., 2016). These limitations have led to a growing interest in using three-dimensional (3D) cell cultures that are more physiologically relevant than 2D cell cultures for metabolism studies (Sood et al., 2019). Here, we show that KSHV infection alters the host spermidine metabolism in 3D culture. KSHV infection led to increased synthesis of intracellular spermidine, a source of eIF5A hypusination, to enhance the translation of LANA protein for viral episomal maintenance in KSHV-infected cells. Inhibition of spermidine synthesis or eIF5A hypusination reduced the nuclear LANA level, which hampered viral episomal maintenance and ultimately led to increased death of KSHV-transformed cells in vitro and in vivo. Our results suggest the inhibition of spermidine metabolic pathway and eIF5A hypusination as therapeutic approaches for targeting KS and PEL tumors.
RESULTS
High level of spermidine accumulation in KSHV-infected cells
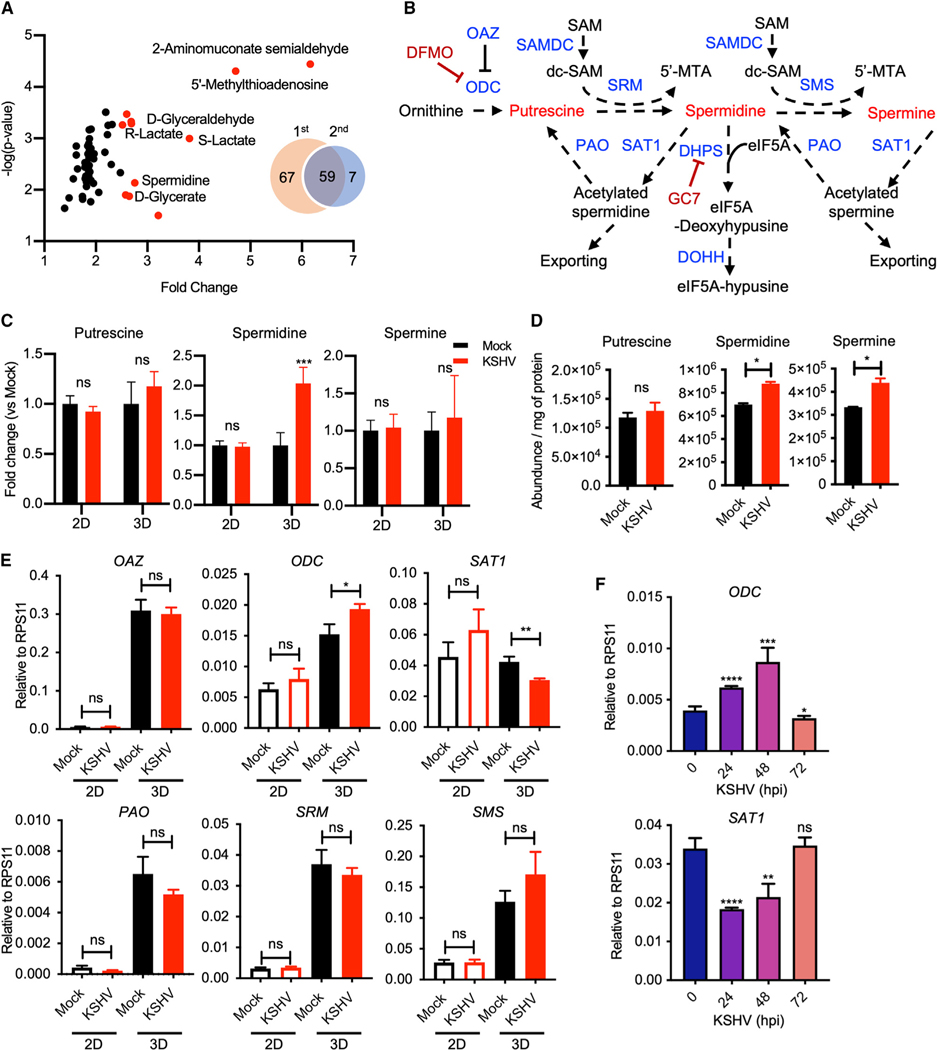
While we have previously tested KSHV-mediated metabolic alterations in a 3D spheroid culture model with Matrigel (Choi et al., 2020; Herrmann et al., 2014), we performed an additional round of metabolomic screening to identify key metabolites influenced by KSHV infection in 3D conditions. Two independent metabolomic analyses revealed 59 common metabolites that were induced in KSHV-infected cells compared with mock-infected cells (Figure 1A and Table S1). Highly upregulated metabolites (>2.5-fold) included those that are linked to the pentose phosphate pathway (D-glyceraldehyde), tryptophan pathway (2-aminomuconate semialdehyde), glycolysis (S-lactate and R-lactate), and polyamine pathway (spermidine and 5′-methylthioadenosine [5′-MTA]). In the eukaryotic polyamine (putrescine, spermidine, and spermine) synthesis pathway, decarboxylated S-adenosylmethionine (dc-SAM) donates an aminopropyl group to putrescine and spermidine to generate spermidine and spermine, respectively (Pegg, 2009; Zappia et al., 1972) (Figure 1B). When KSHV-infected telomerase-immortalized microvascular endothelial (TIME) cells were cultured in 2D or 3D culture conditions, only spermidine out of the three polyamines was highly accumulated in 3D culture, but not in 2D culture (Figure 1C). In concordance with the previous report that polyamines, especially spermidine, increase upon KSHV de novo infection (Delgado et al., 2012), we also detected the accumulation of intracellular spermidine and spermine in TIME cells at 48 h post infection (hpi) with KSHV in 2D culture (Figure 1D).
Figure 1. High level of spermidine accumulation in KSHV-infected cells.
(A) Volcano plot of metabolomic analysis of KSHV-infected 3D TIME cell culture commonly detected from two independent analyses. Venn diagram illustrates thenumber of differentially detected metabolites with p value <0.05 in KSHV-infected 3D culture TIME cells in comparison with mock in two independent metabolomic screenings. Fifty-nine metabolites overlap between the two independent screenings. Red dots represent metabolites upregulated more than 2.5-fold. y axis denotes log10 p values, and x axis shows fold-change values.
(B) The polyamine metabolism pathway. ODC, ornithine decarboxylase; OAZ, ornithine decarboxylase antizyme; SAMDC, S-adenosylmethionine (SAM) decarboxylase; dc-SAM, decarboxylated SAM; 5′-MTA, 5′-methylthioadenosine; SRM, spermidine synthase; SMS, spermine synthase; PAO, polyamine oxidase; SAT1, spermidine/spermine acetyltransferase 1; DHPS, deoxyhypusine synthase; DOHH, deoxyhypusine hydroxylase; DFMO, 2-difluoromethylornithine; GC7, N1-guanyl-1,7-diaminoheptane.
(C and D) Intracellular polyamine levels of mock- and KSHV-infected TIME cells in 2D and 3D culture. (C) Graph represents fold change in comparison with levels in mock-infected cells. Two-way ANOVA with Sidak’s multiple comparisons test was performed to assess the significance. ***p = 0.0003; ns, not significant (biological replicates: 4; technical replicates: 2–3). Error bars represent SEM. (D) Intracellular polyamines in KSHV de novo infected TIME cells harvested at 48 h post infection. *p < 0.05 using Student’s t test; ns, not significant (biological replicates: 2; technical replicates: 2–3). Error bars represent SEM.
(E) qRT-PCR analysis of transcript levels of polyamine metabolism-related enzymes was performed with total RNAs purified from mock- and KSHV-infected TIME cells in 2D (biological replicates: 3; technical replicates: 2–3) and 3D (biological replicates: 4; technical replicates: 3–4) culture. *p = 0.0393, **p = 0.0035; ns, not significant using Student’s t test. Error bars represent SEM.
(F) qRT-PCR analysis of ODC and SAT1 in KSHV de novo infected TIME cells for the indicated times (biological replicates: 2; technical replicates: 2–3). *p < 0.05, **p = 0.0012, ***p = 0.0005, ****p < 0.0001; ns, not significant using Student’s t test. Error bars represent SEM.
To further investigate the alteration of the polyamine pathway induced by KSHV, mRNA transcripts of key enzymes involved in the polyamine metabolism were examined from mock- or KSHV-infected TIME cells in 2D or 3D culture conditions. KSHV-infected TIME cells exhibited the upregulation of ODC expression and the downregulation of SAT1 expression compared with mock-infected cells in 3D culture, but not in 2D culture (Figure 1E). On the other hand, gene expressions of other polyamine metabolizing enzyme genes, ornithine decarboxylase antizyme (OAZ), polyamine oxidase (PAO), spermidine synthase (SRM), and spermine synthase (SMS), showed no difference between mock- and KSHV-infected TIME cells in either 2D or 3D culture. Interestingly, the kinetics of ODC expression increased at 24 h of KSHV de novo infection in 2D culture, reaching a peak at 48 hpi and subsiding to the basal level at 72 hpi, while SAT1 expression decreased at 24 and 48 hpi and returned to the basal level at 72 hpi (Figure 1F). ODC is one of the rate-limiting enzymes in the polyamine biosynthesis pathway that produces putrescine, while SAT1 is the rate-limiting enzyme of the polyamine catabolism pathway that induces the acetylation and secretion of polyamines (Figure 1B). This indicates that KSHV regulates the biosynthesis and catabolism of polyamine to increase the intracellular polyamine metabolites in 3D persistent infection culture conditions or 2D de novo infection culture conditions.
KSHV-induced spermidine synthesis leads to the increase of eIF5A hypusination
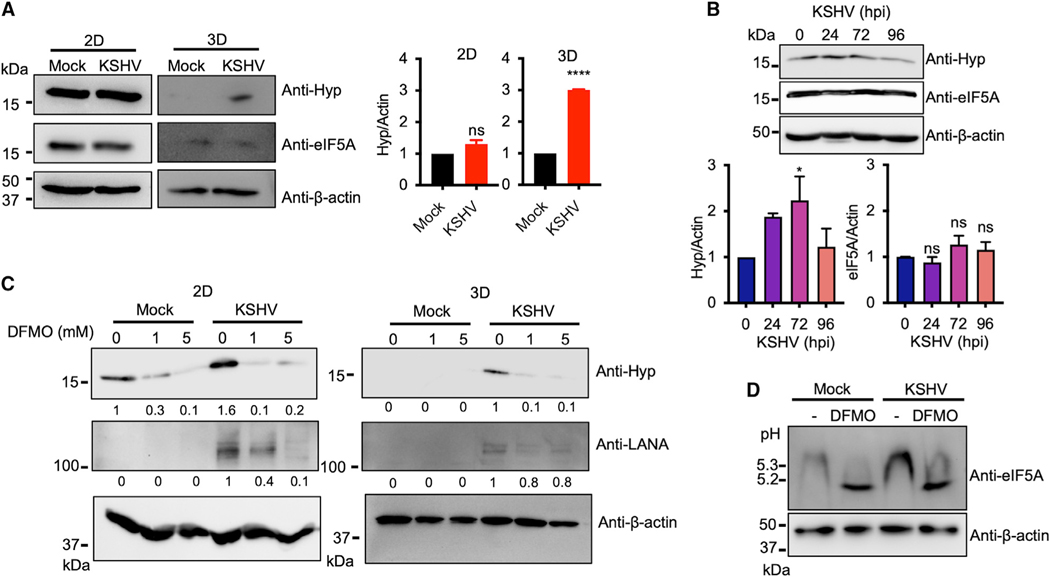
Spermidine acts as a substrate for eIF5A hypusination, which promotes translation of mRNAs encoding clusters of consecutive proline residues that cause ribosome stalling (Gutierrez et al., 2013; Park et al., 1981). As KSHV infection increased intracellular spermidine, we further examined the degree of eIF5A hypusination upon KSHV infection in2Dor 3Dcultureconditions using a hypusine-specific antibody. The hypusination level of eIF5A was noticeably increased in KSHV-infected TIME cells in 3D culture conditions compared with mock-infected TIME cells, while remaining at comparable levels in mock- or KSHV-infected TIME cells in 2D culture conditions (Figure 2A). Moreover, consistent with the increased spermidine level during KSHV de novo infection (Figure 1D), hypusinated eIF5A level was gradually increased until 72 hpi and subsided back to basal level at 96 h of KSHV de novo infection (Figure 2B). Treatment of 2-difluoromethylornithine (DFMO), an inhibitor of ODC (Fozard et al., 1980), significantly reduced the level of eIF5A hypusination in KSHV-infected TIME cells in both 2D and 3D culture conditions, indicating that polyamine biosynthesis directly contributes to the level of eIF5A hypusination upon KSHV infection (Figure 2C). As hypusinated eIF5A has a different isoelectric point from the unhypusinated form, isoelectric focusing gel with anti-eIF5A immunoblotting revealed two distinct bands, the upper band corresponding to the hypusinated eIF5A and the lower band to the unhypusinated eIF5A (Figure 2D). Compared with mock infection, KSHV infection led to an increased level of the upper hypusinated eIF5A band that was shifted to the lower unhypusinated eIF5A band upon DFMO treatment (Figure 2D). These data indicate that KSHV-induced spermidine synthesis results in an increase of eIF5A hypusination.
Figure 2. KSHV-induced spermidine synthesis leads to the increase of eIF5A hypusination.
(A) Immunoblotting analysis of whole-cell lysates from mock- and KSHV-infected TIME cells in 2D (biological replicates: 3; technical replicates: 2–3) and 3D(biological replicates: 2; technical replicates: 2–3) culture. Protein expression was analyzed using anti-hypusine (Hyp), anti-eIF5A, and anti-β-actin antibodies. β-Actin was used as a loading control. Right: representative graph of quantification of hypusination protein level normalized onto β-actin. ****p < 0.0001; ns, not significant using Student’s t test. Error bars represent SEM.
(B) Top: immunoblotting analysis of whole-cell lysates from KSHV de novo infected TIME cells for the indicated times. Protein expression was analyzed using anti-hypusine, anti-eIF5A, and anti-β-actin antibodies. β-Actin was used as a loading control. Bottom: representative graph of the quantification of hypusination (Hyp) and eIF5A levels normalized by β-actin (biological replicates: 3; technical replicates: 2–3). *p = 0.0338; ns, not significant using Student’s t test. Error bars represent SEM.
(C) Immunoblotting analysis of whole-cell lysates from mock- and KSHV-infected TIME cells in 2D or 3D culture. TIME cells were cultured in 2D monolayer and infected with mock or KSHV for 4 h, followed by treatment with 0, 1, or 5 mM DFMO for 48 h (biological replicates: 3; technical replicates: 2–3). Mock- or KSHV-infected TIME cells were cultured in 3D spheroid with 4% Matrigel for 2 weeks, followed by treatment with 0, 1, or 5 mM DFMO for 48 h (biological replicates: 2; technical replicates: 2–3). Whole-cell lysates were probed with anti-hypusine, anti-LANA, and anti-β-actin antibodies by immunoblotting. Hypusination and LANA levels were quantified using Image Lab and normalized to β-actin.
(D) TIME cells were infected with mock or KSHV for 4 h, followed by treatment with 0 mM or 5 mM DFMO for 48 h. Whole-cell lysates were separated in an isoelectric focusing gel (pH 3–10) and probed with anti-eIF5A. Whole-cell lysates separated in SDS-PAGE were probed with anti-β-actin for loading control (biological replicates: 2; technical replicates: 2–3).
eIF5A hypusination is required for efficient KSHV LANA expression
The LANA plays a central role in KSHV episomal tethering and comprises an N-terminal proline-rich region with a total of 18 consecutive proline sequences, a central region with repetitive sequences, and a C-terminal region containing a DNA-binding domain (Figure 3A). Known as a poor donor and acceptor for peptide bond formation in the ribosome during translation, proline often causes ribosome stalling, which requires the hypusinated eIF5A to resume translation (Melnikov et al., 2016). Indeed, the level of LANA expression was drastically reduced in KSHV-infected TIME cells upon DFMO treatment consistently with the decreased eIF5A hypusination (Figure 2C). To assess the effect of eIF5A hypusination on LANA expression, KSHV-infected TIME cells were treated with either DFMO or N1-guanyl-1,7-diaminoheptane (GC7), a DHPS inhibitor (Figure 1B). It showed that either GC7 or DFMO treatment markedly suppressed eIF5A hypusination as well as LANA expression, which was effectively recovered by exogenous spermidine supplementation (Figure 3B). The LANA mRNA level was not affected by treatment of DFMO, GC7, or spermidine supplement (Figure 3C). Consistent with the reduced LANA expression, the number of LANA nuclear dots representing KSHV episome was reduced upon DFMO or GC7 treatment and recovered by spermidine supplementation in either 2D monolayer culture (Figure S1) or 3D spheroid culture of KSHV-infected TIME cells (Figure 3D). Furthermore, LANA expression and eIF5A hypusination of KSHV-infected PEL BCBL-1 cells were also reduced upon DFMO or GC7 treatment (Figure S2A), whereas the LANA mRNA level was not affected (Figure S2B). Correspondingly, LANA nuclear dots were significantly reduced by DFMO or GC7 treatment in BCBL-1 cells (Figure S2C). In addition, expression of viral interferon regulatory factor 3 (vIRF3) latent protein was slightly reduced upon DFMO or GC7 treatment (Figure S2A). Overall, these results suggest that efficient translation of LANA in KSHV-infected endothelial cells and PEL cells largely depends on spermidine-mediated eIF5A hypusination.
Figure 3. eIF5A hypusination is required for KSHV LANA protein expression.
(A) Schematic of LANA. LANA contains a nuclear localization sequence (NLS), proline-rich region, acidic repeat region, and DNA-binding domain. The proline-rich region (N), acidic repeat region (A), and DNA-binding domain (C) used in (F) are highlighted in boxes. (B and C) KSHV-infected TIME cells were treated with 1 mM DFMO or 5 μM GC7, with or without an addition of 20 μM spermidine.
(B and C) Whole-cell lysates were prepared at day 4 post infection and probed for anti-LANA, anti-hypusine, and anti-β-actin antibodies. (B) LANA levels were quantified using Image Lab and normalized to β-actin (biological replicates: 4; technical replicates: 2–3). (C) Extracted RNA were subjected to qPCR analysis for measuring LANA mRNA expression (biological replicates: 2; technical replicates: 2–3). ns, not significant. Error bars represent SEM.
(D) KSHV-infected TIME cells were cultured in 3D spheroid with 4% Matrigel for 2 weeks and treated with 2 mM DFMO or 10 μM GC7, with or without the supplementation of 40 μM spermidine for 5 days, followed by immunofluorescence analysis with anti-LANA antibody (red) and Hoechst 33342 (blue). Images of an entire z stack of 3D spheroid were merged and displayed. Scale bar, 10 μm. Right; graph of the average number of LANA dots per cell (biological replicates: 3).***p < 0.0005, ****p < 0.0001 using one-way ANOVA with Tukey’s multiple comparison test. Error bars represent SEM.
(E) MCF10A cells overexpressing LANA, vIRF3, and EBNA1 were treated with 0, 10, or 50 μM GC7 for 3 days. Protein expression was analyzed using anti-LANA, anti-vIRF3, anti-EBNA1, anti-hypusine, and anti-β-actin antibodies. β-actin was used as a loading control. Each representative graph of quantification of LANA, vIRF3, and EBNA1 level obtained normalized onto β-actin (biological replicates: 3–4; technical replicates: 1–2). **p = 0.0029, ***p = 0.0006; ns, not significant using one-way ANOVA with Dunnett’s multiple comparison test. Error bars represent SEM.
(F) MCF10A cells expressing FLAG-tagged N-terminal domain (N), acidic repeat region (A), or C-terminal domain (C) of LANA were treated with 0, 10, or 50 μM GC7 for 3 days. Top: amino acid sequence of each construct. Consecutive proline sequences are labeled in red. Protein expression was analyzed using anti-FLAG, anti-hypusine, and anti-β-actin antibodies. β-actin was used as a loading control. Each domain of LANA levels was quantified using Image Lab and normalized to β-actin (biological replicates: 2; technical replicates: 2–3).
Hypusinated eIF5A alleviates ribosome pausing at a sequence combination of glycine and aspartic acid besides proline (Pelechano and Alepuz, 2017; Schuller et al., 2017). To further validate the dependency of LANA expression on eIF5A hypusination, we generated MCF10A human breast epithelial cells stably expressing LANA, vIRF3, or Epstein-Barr virus (EBV) EBNA1, a LANA homolog that contains a five proline repeat sequence as well as extensive consecutive glycinealanine sequences. Treatment of GC7 readily downregulated LANA expression in MCF10A cells, whereas little or no effect on vIRF3 and EBNA1 expression was observed (Figure 3E). To exclude the potential effect of GC7 treatment on mRNA transport, in vitro transcribed 5′-capped and polyadenylated mRNA of LANA, vIRF3, or EBNA1 was transfected into HEK-293T cells (Figure S3A) and its de novo expression was quantified. Consistently, LANA expression from in vitro transcribed mRNA was markedly reduced upon GC7 treatment, whereas only a marginal reduction of EBNA1 or vIRF3 expression was detected under the same conditions (Figure S3B).
The frequency of polyproline motifs negatively correlates with translational efficiency (Qi et al., 2018). EBNA1 and vIRF3 have five and four consecutive proline sequences, respectively, while LANA contains 18 consecutive proline sequences, suggesting that the higher frequency of polyproline motifs causes the more robust translational pause that requires hypusinated eIF5A for efficient translation. To further assess the dependency of the LANA proline-rich region to hypusinated eIF5A, we generated MCF10A cells stably expressing the N-terminal proline-rich region (N), a central acidic repeat region (A), or C-terminal DNA-binding region (C) of LANA (Figures 3A and 3F). GC7 treatment noticeably reduced expression of the N region and slightly diminished expression of the A region, but did not alter expression of the C region (Figure 3F). These results suggest that the N-terminal proline-rich domain of LANA is primarily responsible for ribosome stalling and requires eIF5A hypusination for efficient translation.
DFMO and GC7 suppress viral episomal maintenance and induce tumor cell death in vitro and in vivo
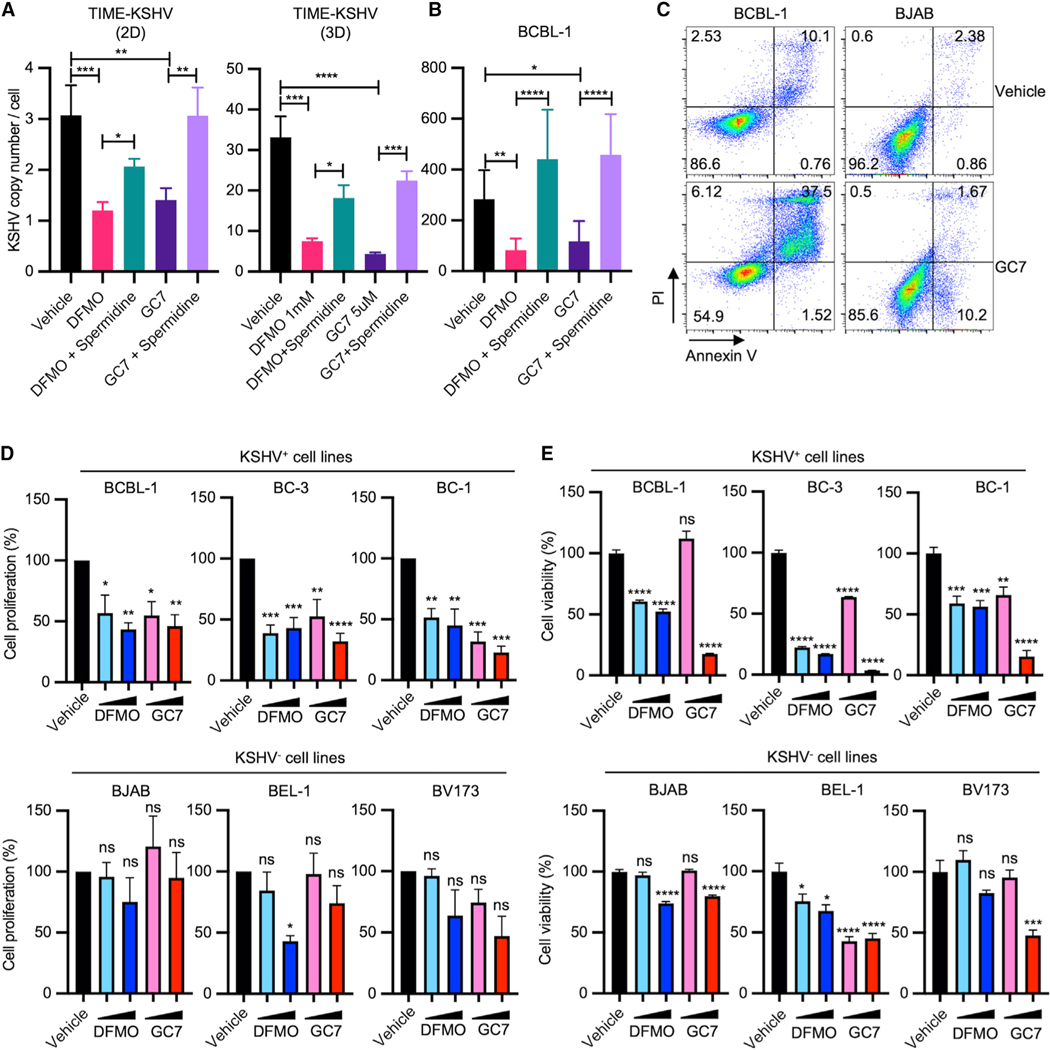
LANA mediates viral episomal maintenance by tethering KSHV genomes to the host mitotic chromosomes to segregate its genomes to the daughter cell nuclei. To explore the effect of polyamine synthesis inhibitor (DFMO) and hypusination inhibitor (GC7) on viral episomal maintenance, KSHV-infected TIME or BCBL-1 cells were treated with DFMO or GC7, followed by measurement of KSHV genome copy number by qPCR. Treatment of DFMO or GC7 led to a marked reduction of viral genome copy numbers in 2D- or 3D-cultured KSHV-infected TIME cells (Figure 4A) and 2D-cultured BCBL-1 cells (Figure 4B), but the reduced KSHV genome copy numbers were restored by exogenous spermidine supplementation. Furthermore, GC7 treatment resulted in robust apoptotic cell death of BCBL-1 cells, whereas it led to a minor effect on KSHV-negative Burkitt’s lymphoma BJAB cells (Figure 4C). To further explore whether LANA-mediated viral episomal maintenance is critical for cell viability, we examined the proliferation rate and viability of KSHV-positive PEL cells (BCBL-1, BC-1, and BC-3) and KSHV-negative lymphoma or leukemia cells (BJAB, BEL-1, and BV173) (Tang et al., 2004). DFMO or GC7 treatment also limited the proliferation of KSHV-infected BCBL-1, BC-1, and BC-3 PEL cells, whereas KSHV-negative BJAB, BEL-1, and BV173 cells did not show significant change in their proliferation rates (Figure 4D). In addition, we observed drastic reduction of KSHV-positive PEL cell viability upon DFMO or GC7 treatment in a dose-dependent manner, whereas KSHV-negative cells showed only modest reduction of viability at high-dose treatments of DFMO or GC7 (Figure 4E). Collectively, these results indicate that inhibition of the spermidine-hypusination axis reduces LANA expression and activity in viral persistence, which ultimately leads to retarded proliferation and enhanced death of KSHV-infected cells.
Figure 4. DFMO and GC7 restrain KSHV persistence and induce cell death in KSHV-infected cells.
(A and B) (A) KSHV-infected TIME cells in 2D or 3D culture and (B) BCBL-1 cells in 2D culture were treated with 1 mM DFMO or 5 μM GC7, with or without the supplementation of 20 μM (2D culture) or 40 μM (3D culture) spermidine for 5 days. Copy numbers of intracellular KSHV genomes were quantified by qPCR from purified genomic DNA (biological replicates: 3–4; technical replicates: 3–4). Each sample was normalized to cellular b-globin. *p < 0.05, **p < 0.005, ***p = 0.002, ****p < 0.0001 using one-way ANOVA with Tukey’s multiple comparison test. Error bars represent SEM.
(C) BCBL-1 and BJAB cells were treated with or without 10 μM GC7 for 4 days. Apoptosis was assessed by flow cytometry by annexin V and propidium iodide (PI) staining (biological replicates: 2–3). Numbers indicate the percentage of cells in each quadrant.
(D and E) (D) Viability and (E) proliferation of BJAB, BCBL-1, BC-3, BC-1, or JSC-1 cells upon DFMO (0.5 and 1 mM) or GC7 (5 and 10 μM) treatment for 5 days were measured by cell counting and CellTiter-Glo, respectively (biological replicates: 3). *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001; ns, not significant using one-way ANOVA with Dunnett’s multiple comparison test. Error bars represent SEM.
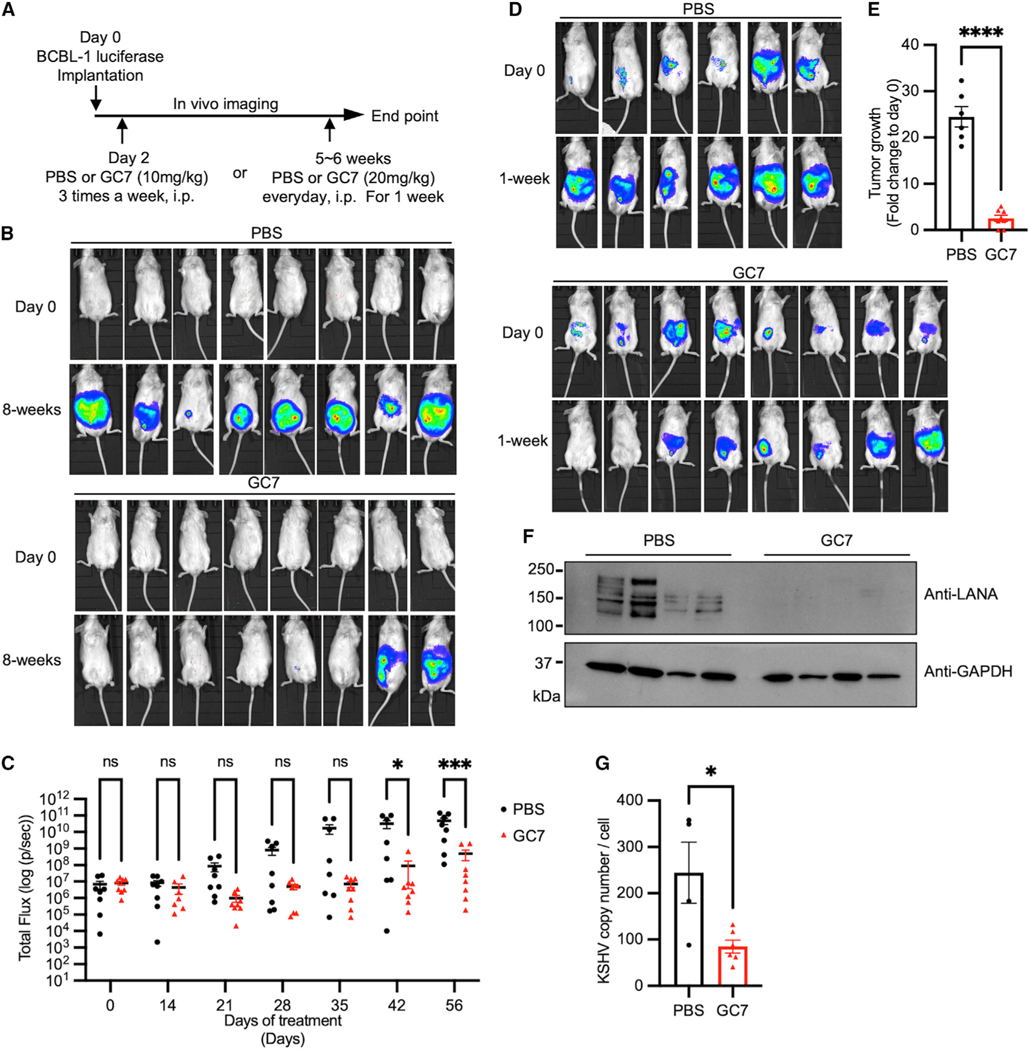
To evaluate the efficacy of GC7 as a treatment for KSHV-positive PELs or other lymphoma cells, NOD/SCID xenograft mice were intraperitoneally injected with KSHV-positive BCBL-1-luciferase cells, EBV-transformed LCL352-luciferase cells, or KSHV-negative/EBV-negative BJAB-luciferase lymphoma cells as previously described (Lee et al., 2011). At day 2 post engraftment, NOD/SCID xenograft mice were treated daily with either PBS or GC7 via intraperitoneal injection (Figures 5A, S4A, and S5A). All BCBL-1 engrafted PBS-treated mice (8 out of 8) started to display evident distention and ascites in the peritoneal cavity with markedly increased luminescence signal around 5 weeks post engraftment (Figures 5B and 5C). GC7 treatment effectively reduced tumor development in most BCBL-1-engrafted mice (tumor formation in 2 out of 8 mice) (Figures 5B and 5C). In contrast, it showed weak effect on tumor development in EBV-transformed LCL352-engrafted mice (tumor formation in 5 out of 8 mice) (Figures S4B and S4C) and no effect on tumor development in KSHV-negative/EBV-negative BJAB-engrafted mice (tumor formation in 13 out of 13 mice) (Figures S5B and S5C).
Figure 5. Anti-tumor activity of hypusination inhibitor against PEL growth in vivo.
(A) NOD/SCID mice were intraperitoneally (i.p.) engrafted with 5 × 106 BCBL-1-luciferase cells and imaged using the IVIS imaging system for bioluminescence signal intensity in the whole body.
(B) Two days post engraftment, mice were injected (i.p.) with PBS or GC7 three times a week for 8 weeks. Engrafted mice are shown before or after treatment.
(C) Quantification of in vivo bioluminescence signal of the animals treated with PBS or GC7 for 8-week treatment period (n = 8 per group). Two-way ANOVA with Sidak’s multiple comparisons test was performed to assess the significance. *p = 0.0195, ***p = 0.0001 ns, not significant.
(D) BCBL-1-luciferase tumor-bearing mice were injected (i.p.) with PBS or GC7 daily for a week. Mice are shown before or after treatment.
(E) Tumor growth for each mouse from (D) was measured by in vivo bioluminescence signal measurement. Fold change of total flux for each mouse was graphed as a bar (n = 6–8 per each group). ****p < 0.0001 using Student’s t test. Error bars represent SEM.
(F and G) Tumor cells from the ascites of mice (D) were isolated and analyzed for (F) immunoblotting to show LANA and GAPDH (loading control) expression or (G) qPCR to measure intracellular KSHV genome copy numbers. *p = 0.0198 using Student’s t test. Error bars represent SEM.
To examine whether GC7 treatment induces the regression of KSHV-positive PEL-induced tumors or EBV LCL-induced tumors, 5- to 6-week post-engraftment mice were randomly separated into two groups for PBS or GC7 treatment. After 1 week of treatment, BCBL-1 tumor-bearing mice (8 out of 8) showed ~25-fold increases of luminescent signals upon PBS treatment (Figures 5D and 5E). In contrast, a majority of BCBL-1 tumor-bearing mice (6 out of 8) showed minor or no increases of luminescent signals upon GC7 treatment, and two mice displayed an undetectable level of luminescence signals (Figures 5D and 5E). On the other hand, no detectable regression of EBV-positive LCL352-induced tumors was observed after GC7 treatment (Figures S4D and S4E).
To explore the mechanism underlying GC7-induced tumor regression, we collected ascites and solid tumors from BCBL-1 engrafted mice treated with PBS and residual tumor cells from BCBL-1 engrafted mice treated with GC7. GC7 treatment induced considerable reduction of the KSHV LANA expression in ascites from BCBL-1 engrafted mice compared with PBS treatment in ascites and solid tumors from BCBL-1 engrafted mice (Figure 5F). In contrast, the EBV EBNA1 expression of LCL352-induced tumors was not detectably affected by GC7 treatment (Figure S4F). Consequently, KSHV genome copy numbers of BCBL-1-induced tumors were reduced to 3-fold upon GC7 treatment compared with that upon PBS treatment (Figure 5G). In contrast, EBV genome copy numbers were not altered by GC7 treatment in LCL352-induced tumors (Figure S4G). Collectively, these results indicate that GC7 treatment shows a specific effect on KSHV LANA expression.
DISCUSSION
Oncogenic viruses markedly alter the host metabolism to self-fulfill the need for metabolic intermediates required for viral tumorigenesis (Mesri et al., 2014). Understanding how oncogenic viruses exploit specific metabolic pathways is critical for therapeutic target identification of virus-associated cancer. However, the conventional 2D culture condition does not fully mimic the in vivo metabolic niche, as the tissue architecture and cell-cell contacts are absent in 2D culture systems. Here, we demonstrate that KSHV infection increases spermidine metabolites and eIF5A hypusination in 3D culture as well as in the 2D de novo infection condition. Furthermore, we determine that hypusinated eIF5A alleviates the translational restriction of LANA expression in KSHV-infected cells. Our study reveals that the spermidine-hypusination signaling axis is essential for LANA expression and viral episomal maintenance of KSHV-infected cells. Conversely, suppression of the spermidine-hypusination signaling axis leads to a decrease of LANA expression and viral episomal maintenance, followed by pronounced apoptotic death of KSHV-associated tumor cells.
Polyamines have been implicated to play a role in various stages of virus infection, including viral replication and assembly (Mounce et al., 2017). Polyamines, found in the nucleocapsid of herpesvirus virions are presumed to facilitate virion assembly of human cytomegalovirus and herpes simplex virus 1, where depletion of the polyamine effectively diminished virus production (Gibson et al., 1984; Greco et al., 2005; Tyms and Williamson, 1982). In addition, inhibiting polyamine synthesis also reduces the replication of Ebola virus (EBOV) (Olsen et al., 2016). Recent work has suggested that type I interferon induces SAT1, a polyamine catabolic enzyme, as an anti-viral defense to decrease polyamines, as Chikungunya virus and Zika virus require polyamines for transcription and translation (Mounce et al., 2016). Moreover, eIF5A hypusination also controls EBOV gene expression (Olsen et al., 2018). These previous findings strongly indicate that polyamines are crucial mediators of the virus life cycle.
Polyamine levels are shown to be elevated in various types of cancer. In colon cancer, APC tumor-suppressor gene and KRAS oncogene have been shown to regulate expression of the ODC and SAT1 genes. For instance, a high ODC level, which leads to increased polyamine biosynthesis, and low SAT1 level, which reduces polyamine export, mutually induce the accumulation of polyamine and contribute to tumorigenesis in colon cancer (Ignatenko et al., 2011; Li et al., 2020). In a mouse colon cancer model, tumorigenesis is reduced by a combinatory regimen of non-steroidal anti-inflammatory drug treatments that increases SAT1-mediated cellular export of polyamines, together with DFMO treatment that decreases ODC-mediated polyamine synthesis (Jacoby et al., 2000). EBV-positive Burkitt’s lymphoma also exhibits reduced SAT1 gene expression and increased polyamine, which has a favorable effect on the tumorigenic feature of Burkitt’s lymphoma (Shi et al., 2013). In concurrence with these reports, we demonstrate that the regulation of ODC and SAT1 transcription is the key mechanism in KSHV-mediated alteration of polyamine metabolism for viral persistence and tumorigenesis. PEL cell death induced by DFMO treatment also highlights the importance of the polyamine pathway in KSHV tumorigenesis. Myc oncoprotein is one of the activators of ODC transcription, promoting polyamine synthesis and proliferation of many cancer cells. During latency, KSHV has been known to stabilize Myc protein through latent proteins, such as vIRF3 and LANA (Baresova et al., 2012; Liu et al., 2007). KSHV-mediated stabilization of Myc proteins may positively regulate the polyamine anabolic pathway, enhancing eIF5A hypusination to increase LANA expression for viral persistence.
Cells cultured in 3D conditions have been shown not only to manifest different gene expression profiles from those cultured in 2D conditions but also to highly recapitulate cells grown in an in vivo setting (Birgersdotter et al., 2005; Ghosh et al., 2005). 3D cultured cells generally upregulate expressions of multiple genes associated with stress response, inflammation, redox signaling, hypoxia, and angiogenesis (Jensen and Teng, 2020). We also found that basal expressions of the polyamine metabolism enzyme genes (OAZ, SRM, and SMS) were increased in 3D culture. This suggests that the stressful environments in 3D culture, including limited oxygen and nutrients and accumulated metabolic waste, may play roles in regulating these gene expressions. Intriguingly, functional OAZ enzyme expression is regulated by intracellular polyamine level (Coffino, 2001). At a low level of polyamine, the translation of OAZ is shortened at a premature stop codon, causing non-functional protein synthesis. In contrast, a high level of polyamine introduces ribosomal frameshift that leads to skipping of the premature stop codon and inducing functional OAZ expression (Matsufuji et al., 1995; Rom and Kahana, 1994). As the basal level of hypusination in 3D culture was lower than that in 2D culture (Figures 2A and 2C), the limited source of polyamines in 3D culture may be one of many factors that cause differences in KSHV-induced metabolic changes in 3D culture versus 2D culture.
Unlike primary KS tumors in patients, KSHV episomes are rapidly lost in in vitro cell-culture systems. More recently, this incongruity has been addressed by a study (Dubich et al., 2021) reporting that compared with 2D culture, KSHV genomes explicitly increase during de novo infection and are stably maintained under 3D culture. This suggests that continuous de novo infection in 3D culture may be critical for KSHV persistence. Intriguingly, our 3D spheroid model exhibited increased KSHV genome copy numbers per cell compared with the 2D culture system (Figure 4A). This suggests that KSHV induces spermidine biosynthesis and eIF5A hypusination to increase LANA expression, thereby achieving LANA-mediated episomal maintenance. In addition to LANA, it is highly plausible that other host proteins in 3D cultured KSHV-infected cells may also be targets of eIF5A hypusination to concertedly contribute toward viral persistence and tumorigenesis. In fact, hypusinated eIF5A enhances the translation of proteins that are involved in ECM organization and collagen metabolism for tumor progression in vivo and 3D spheroid formation (Henke et al., 2019; Kauppila et al., 1998; Pelechano and Alepuz, 2017). This suggests that dysregulation of the polyamine metabolism and eIF5A hypusination may perturb the integrity of the ECM proteins and collagens in 3D culture or in vivo infection. Future study of host or viral protein expressions that are altered by KSHV-induced eIF5A hypusination will provide mechanistic insights into the spermidine-hypusination signal-mediated regulation of the viral life cycle.
Among the three polyamines, only spermidine donates its aminobutyl group to eIF5A, which consequently results in hypusination and activation of eIF5A (Saini et al., 2009). Our findings reveal that the increased intracellular level of spermidine in 3D KSHV-infected cells or 2D de novo KSHV infection leads to increased eIF5A hypusination. Hypusinated and activated eIF5A, which alleviates ribosome stalling during the synthesis of polyproline motif-containing proteins, subsequently allowed efficient translation of the proline-rich LANA for the maintenance of viral episome. Treatment of PEL cells with GC7 effectively suppressed eIF5A hypusination, LANA translation, viral episomal maintenance, and cell proliferation in a dose-dependent manner. This suggests that KSHV targets the spermidine-hypusination signal pathway for viral episomal maintenance as well as for virus-induced tumorigenesis. Our study provides not only molecular details underlying how KSHV exploits the intracellular spermidine-hypusination signal pathway but also an invaluable insight into a therapeutic target to treat KSHV-associated cancer.
Limitations of the study
We have found that KSHV infection regulates the transcriptions of polyamine metabolism-related genes and subsequently leads to increased spermidine production. However, detailed mechanisms of KSHV-mediated regulation of polyamine metabolic gene expressions remain unknown. In addition, while we have discovered a specific effect of eIF5A hypusination on the KSHV LANA translation, additional study is needed to investigate broad effects of eIF5A hypusination on the translational controls of host and viral proteins.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jae U. Jung (JUNGJ@ccf.org).
Materials availability
All unique materials generated during this study will be made available upon request to the lead contact.
Data and code availability
The raw metabolomics data reported in this study are available in the MetaboLights database (https://www.ebi.ac.uk/metabolights/index) under accession numbers MTBLS5218.
The metabolome processed data reported in this study are supplied as Table S1. All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
Cells were maintained at 37°C in a humidified incubator with 5% CO2. HEK-293T and iSLK-BAC16 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Seradigm) and 1% penicillin/streptomycin (Gibco). iSLK-BAC16 cells were cultured in the presence of 1 μg/mL of puromycin, 250 μg/mL of G418, and 1 μg/mL of hygromycin. TIME cells were cultured in VascuLife VEGF Endothelial Medium (LIFELINE). MCF10A cells were cultured in DMEM-F12 supplemented with 5% horse serum (Thermofisher), EGF 20 ng/mL (Peprotech), insulin 10 μg/mL (Sigma), hydrocortisone 0.5 mg/mL (Sigma), cholera toxin 100 ng/mL (Sigma) and 1% penicillin/streptomycin (Gibco). The PEL cell lines (BCBL-1, BC-1, and BC-3) and KSHV-negative control cells (BJAB, BEL-1, BV173, and LCL352-mCherry-luciferase) were cultured in RPMI 1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin. The BCBL-1 luciferase and BJAB luciferase cell line were maintained in RPMI 1640 medium with 50 μg/mL and 100 μg/mL of hygromycin, respectively. To generate 3D spheroid, TIME cells with 4% Matrigel (Corning) were seeded at 5 × 104 cells per 6-well ultralow attachment plates (Corning). Cultures were fed every 3–4 days with assay medium.
Animal models
Female NOD/SCID (5 weeks old) were purchased from Jackson. After 1 week adaptation, BCBL-1, LCL352, and BJAB luciferase cells (5 × 106 cells) were intraperitoneally (i.p) injected and 2 days post cells engraftment, GC7 (10 mg/kg) was intraperitoneally injected 3 times a week to observe effect of GC7 on tumor establishment. To monitor tumor regression, GC7 (20 mg/kg) was intraperitoneally injected daily for a week after mice developed cancer cells with evidently increased luminescence. For luciferase imaging, mice will be anesthetized with 2% isoflurane in 100% oxygen. D-luciferin firefly potassium salt (Glodbio) will be injected intraperitoneally (150 mg/kg), and mice will subsequently be imaged in the IVIS system (PerkinElmer) while sedated. A region of interest (ROI) was manually selected over the signal intensity. Data are presented as average radiance. General behavior were recorded every other day to ensure animal health was maintained. All experiments were approved and done according to the guidelines of the Institutional Animal Care and Use Committee at the Cleveland Clinic.
METHOD DETAILS
KSHV preparation and infection
KSHV virus was prepared from iSLK-BAC16 cells as previously described (Brulois et al., 2012; Myoung and Ganem, 2011). Briefly, 70% confluent iSLK-BAC16 cells were induced with growth medium containing 1 mM sodium butyrate (Sigma) and 1 μg/mL doxycycline (Sigma). 4 days later, culture medium containing virus was collected and cleared by centrifugation and filtering to remove cells and debris. Virus was concentrated by ultracentrifugation for 3 h at 24,000 rpm in SW32 rotor (Beckman Coulter). Virus pellets were resuspended in Dulbecco′s Phosphate Buffered Saline (DPBS) and stored at −80°C. For de novo infection of KSHV, when TIME cells reached 60 to 70% confluence, cells were incubated with KSHV for 4 h at 37°C. After the inoculum was removed, and the cells were washed with DPBS three times, followed by addition of growth medium before being returned to the incubator. For making stably latent infected cell, KSHV infected TIME cells were selected and maintained with 10 μg/mL of hygromycin.
Quantitative RT-qPCR
RNA was extracted using TRIzol reagent (Invitrogen), according to manufacturer’s instructions. Total RNA used for cDNA synthesis using iScript (Bio-Rad), according to manufacturer’s instructions. RT-qPCR using SyberGreen (BioRad) was performed on CFX96 Real-time PCR (Bio-Rad), according to manufacturer’s instructions. Samples were run in triplicate and normalized to the ribosomal protein S11 (rps11). Relative expression was calculated using the ΔΔCT method. Primer sequences are found in Table S2.
Isoelectric focusing
Cells were lysed in low salt hypotonic buffer (10 mM HEPES, pH 7.3, 1.5 mM MgCl2, 20 mM KCl, 500 μM dithiothreitol, 1% Triton X-100) supplemented with protease inhibitor mixture for 15 min on ice. Equal amount of protein extract was resolved in Novex pH 3–10 IEF Protein Gels (ThermoFisher). The transferred onto PVDF membranes for immunoblotting analysis.
Immunoblotting
Cells were lysed in RIPA buffer (50 mM Tris-HCl (pH7.4); 150 mM NaCl; 1% NP-40; 0.25% deoxycholic acid; 1mM EDTA) supplemented with protease inhibitor mixture and quantified by BCA (Pierce). Equal amount of protein extract was resolved on SDS-PAGE gels and transferred onto PVDF membranes. Antibody concentrations were as follows: anti-hypusine, anti-eIF5A, anti-LANA, anti-vIRF3, anti-EBNA1, anti-Flag, anti-β-actin and anti-GAPDH. They were incubated with specific antibodies followed by HRP-conjugated secondary antibodies and Images were developed with ECL reagent (Millipore) and imaged on a Bio-Rad ChemiDoc-Touch.
Metabolite extraction and liquid chromatography-mass Spectrometric analysis (LC-MS)
2D monolayer cultured TIME cells were collected and washed twice with cold PBS and fixed in 80% methanol (Fisher Chemical, LC-MS glade) precooled in dry ice. Cells were detached by scrapping and then transferred to a 2 mL screw cap tube. 3D cultured TIME cells are incubated with Cell Recovery Solution (Corning) for 20 min on ice for depolymerize the Matrigel matrix before fixation. Lysate of cells were quantified by BCA and pelleted by centrifugation (12000 rpm for 10 min at 4°C). The supernatant was transferred into a new 2 mL screw cap tube and driedusing a vacuum concentrator. Dried metabolites were resuspended in acetonitrile/methanol/water(40:40:20).Before applying LC-MS, metabolites were mixed with acetonitrile containing 0.2% folic acid solution and spun down at 12,000 rpm for 10min at 4°C. The supernatant was injected into Agilent Accurate Mass 6230 time of flight (TOF) coupled with Agilent 1290 liquid chromatography (LC) system. Detected ions were deemed metabolites on the basis of unique accurate mass-retention time identifiers for masses exhibiting the expected distribution of accompanying isotopologues. The abundance of metabolites was extracted using Agilent Qualitative Analysis B.07.00 and Profinder B.08.00 software (Agilent Technologies) with a mass tolerance of <0.005 Da.
Immunofluorescence
KSHV infected TIME cells were plated on coverslip and BCBL-1 cells were plated on coverslips coated with Poly-L-lysine solution (Sigma) for 1 h after treatment with or without DFMO, GC7, and spermidine. 3D spheroids of KSHV infected TIME cells are transferred to 1.5 mL tube to proceed staining procedure in tubes. Cells were fixed with 100% methanol for 10 min at −20°C and blocked 10% goat serum in PBS. Fixed cells were stained with anti-LANA (1:500) diluted in 1% BSA-PBS for 2 h. Appropriate fluorescence-conjugated secondary antibodies from goat were incubated for 1 h. DNA was stained with 1 μg/mL Hoechst 33342. Coverslips of 2D cultured KSHV infected TIME cells and BCBL-1 cells were mounted with Fluoromount-G (Thermo Scientific) mounting media. For 3D spheroid of KSHV infected TIME cells, stained spheroids pellets were resuspended in mounting media and transferred to slides. Cells and spheroids were examined under Leica SP8 confocal microscope. Leica LAS-X software was used to merge and stack individual confocal sections of 3D spheroid through the z axis to generate maximum projection images.
mRNA synthesis and transfection
Coding region of LANA, EBNA1, and vIRF3 were inserted to pZMV plasmid that contains a T7 promoter. The capped mRNA was synthesized from corresponding linearized pZMV plasmid using the HiScribe T7 High Yield RNA Synthesis Kit (NEB) and Cleancap reagent (Trilink). After a total incubation for 2 hours at 37°C, the DNA was digested by DNase I treatment and mRNA was purified using RNA Clean & Concentrator-5(Zymo). Polyadenylation was performed by using E.coli Poly(A) Polymerase (NEB).The mRNA was again purified using RNA Clean & Concentrator-5. mRNA concentration was determined on a NanoDrop OneC (Thermo Fisher). mRNA quality was determined by 1% formaldehyde agarose gel. In vitro transcribed mRNA was transfected to HEK-293T cells using TransIT reagent (Mirus).
Measuring viral genome copy numbers
DNA was extracted from cells using a MasterPure DNA purification kit (Lucigen) according to the manufacture’s protocols. To measure viral genome copy number per cell, we used 20 ng of DNA for qPCR with SsoAdvanced Universal SYBR Green Supermix as previously described (Wu et al., 2006). Briefly, ORF11 primers were used to detect KSHV genomes, and BAC16 bacmid was used as a standard. To estimate the number of EBV genome copies, EBNA1 primers were used to detect EBV genomes, and pCXWB- EBNA1 (Addgene) plasmid was used as a standard. To determine genome copy number, β-globin primers were used detect host chromosome, and plasmid containing fragment of β-globin was used for host chromosome standard. Primer sequences are listed in Table S2.
Apoptotic assay and cell viability assay
FITC Annexin V (Biolegend) was used according to the manufacturer’s instruction, and PI solution (Biolegend) was applied at 20 μg/mL. Analysis was performed on singlet cells. The cells were incubated for 15 min in the dark and analyzed with a FACS Celesta flow cytometer (BD Biosciences). To determine cell viability, the CellTiter-Glo Luminescent Cell Viability Assay (Promega) was performed according to the manufacturer’s instruction.
QUANTIFICATION AND STATISTICAL ANALYSIS
Flow cytometry data was analyzed by FlowJo v.10.7.1. The statistical tests were calculated using GraphPad Prism 9. We performed a Student’s t-test or onw-way ANOVA or two-way ANOVA to calculate p values with statistical significance. For each experiment, the number of biological or technical replicates are provided in the figure legends. Details of the specific statistical analysis and p-values are indicated in the figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Rabbit anti-eIF5A | Abcam | Cat# ab32443; RRID: AB_2230904 |
| Rabbit anti-Hypusine | Raghu Mirmira Lab | N/A |
| Rabbit anti-Hypusine | Millipore | Cat# ABS1064; RRID: AB_2631138 |
| Rat anti-LANA | Advanced Biotechnologies | Cat# 13-210-100; RRID: AB_1929223 |
| Mouse anti-vIRF3 | Novus | Cat# CM-A807 |
| Rabbit anti-EBNA | Paul Lieberman Lab | N/A |
| Rabbit anti-Flag | Sigma | Cat# F7425; RRID: AB_439687 |
| Mouse anti-β-actin | Santa Cruz | Cat# sc-47778; RRID: AB_626632 |
| Horse Anti-mouse IgG-HRP | Cell Signaling | Cat# 7076; RRID: AB_330924 |
| Goat Anti-rabbit IgG-HRP | Cell Signaling | Cat# 7074; RRID: AB_2099233 |
| Goat Anti-rat IgG-HRP | Santa Cruz | Cat# sc-2006; RRID: AB_112521 |
| Goat anti-Rat IgG-Alexa Fluor 568 | Thermo Fisher | Cat# A-11077; RRID: AB_2534121 |
| Mouse anti-GAPDH | Santa Cruz | Cat# sc-47724; RRID: AB_627678 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| BAC16 KSHV | Brulois et al., 2012 | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Spermidine | Cayman Chemical | 14918 |
| Doxycycline hyclate | Sigma | D9891 |
| Sodium butyrate | Sigma | 303410 |
| Novex™ pH 3-7 IEF Protein Gels, 1.0 mm, 10-well |
ThermoFisher | EC6645BOX |
| Growth Factor Reduced (GFR) Basement Membrane Matrix, Phenol Red-free (Matrigel) |
Corning | 356231 |
| FITC-Annexin V | Biolegend | 640906 |
| Propidium Iodide | Biolegend | 421301 |
| TRIzol | Thermo Fisher | 15596018 |
| iScript™ cDNA Synthesis Kit | Bio-Rad | 1708891BUN |
| SsoAdvanced Universal SYBR Green Supermix | Bio-Rad | 1725272 |
| GC7 | Millipore | 259545 |
| DFMO | Cayman Chemical | 16899 |
| D-Luciferin, Potassium Salt | Goldbio | LUCK-100 |
| Poly-L-lysine solution | Sigma | P4707 |
| CleanCap AG | Trilink | N-7413-5 |
| E. coli Poly(A) Polymerase | New England Biolabs | M0276 |
| Methanol, LC/MS Grade | Fisher Chemical | A456 |
| Acetonitrile, LC/MS Grade | Fisher Chemical | A955 |
|
| ||
| Critical commercial assays | ||
|
| ||
| CellTiter-Glo 2.0 Assay | Promega | G9241 |
| MasterPure Complete DNA and RNA Purification Kit | Lucigen | MC85200 |
| HiScribe T7 High Yield RNA Synthesis Kit | New England Biolabs | E2040S |
| RNA Clean & Concentrator-5 | Zymo Research | R-1014 |
|
| ||
| Deposited data | ||
|
| ||
| Normalized mass spectrometry metabolomics data | This paper | Table S1 |
| Raw mass spectrometry metabolomics data | This paper | MTBLS5218 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Human: Embryonic kidney 293T cells | ATCC | CRL-3216 |
| Human: iSLK cells | Myoung J et al., 2011 | N/A |
| Human: iSLK-BAC16 cells | Brulois et al., 2012 | N/A |
| Human: BCBL-1 | Donald Ganem Lab | N/A |
| Human: BCBL-1-luciferase | Lee et al., 2011 | N/A |
| Human: BC-1 | ATCC | CRL-2230 |
| Human: BC-3 | ATCC | CRL-2277 |
| Human: BJAB | DSMZ | ACC 757 |
| Human: BEL-1 | Tang et al.,2004 | N/A |
| Human: BV173 | DSMZ | ACC 20 |
| Human: BJAB-luciferase | This paper | N/A |
| Human: LCL352-mCherry-luciferase | Paul M. Lieberman Lab | N/A |
| Human: Endothelial cells immortalized with hTERT (TIME) | ATCC | CRL-4025 |
| Human: Mammary gland epithelial cells (MCF10A) | ATCC | CRL-10317 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: NOD/SCID | The Jackson Laboratory | 001303 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primer for qPCR (See Table S2) | Integrated DNA Technologies | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pCDH-puro-LANA | This paper | N/A |
| pCDH-puro-LANA (N) | This paper | N/A |
| pCDH-puro-LANA (A) | This paper | N/A |
| pCDH-puro-LANA (C) | This paper | N/A |
| pCDH-puro-vIRF3 | This paper | N/A |
| pCDH-puro-EBNA1 | This paper | N/A |
| pCXWB-EBNA1 | Addgene | Plasmid #37624 |
| pZMV plasmid | This paper | N/A |
| pZMV-LANA | This paper | N/A |
| pZMV-vIRF3 | This paper | N/A |
| pZMV-EBNA1 | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Prism 9 | GraphPad Software | N/A |
| Agilent Qualitative Analysis B.07.00 | Agilent Technologies | N/A |
| Profinder B.08.00 | Agilent Technologies | N/A |
| FlowJo | BD | N/A |
Highlights.
KSHV infection induces spermidine synthesis in 3D culture
Increased spermidine level promotes eIF5A hypusination
Hypusinated eIF5A is required for efficient translation of the KSHV LANA protein
Inhibition of eIF5A hypusination suppresses KSHV episome maintenance and oncogenesis
ACKNOWLEDGMENTS
We are grateful to Dr. Raghu Mirmira for the hypusine antibody and Dr. Paul Lieberman for LCL352-mCherry-luciferase cells and EBNA1 antibody. This work was partly supported by: grant nos. CA200422, CA251275, AI140718, AI140705, AI140705S1, AI152190, AI171201, DE023926, DE028521, and KGM9942011, and Betsy B. deWindt Endowment to J.U.J.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111234.
REFERENCES
- Agostinelli E, Marques MP, Calheiros R, Gil FP, Tempera G, Viceconte N, Battaglia V, Grancara S, and Toninello A. (2010). Polyamines: fundamental characters in chemistry and biology. Amino Acids 38, 393–403. 10.1007/s00726-009-0396-7. [DOI] [PubMed] [Google Scholar]
- Ballestas ME, Chatis PA, and Kaye KM (1999). Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284, 641–644. 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- Baresova P, Pitha PM, and Lubyova B. (2012). Kaposi sarcoma-associated herpesvirus vIRF-3 protein binds to F-box of Skp2 protein and acts as a regulator of c-Myc protein function and stability. J. Biol. Chem 287, 16199–16208. 10.1074/jbc.M111.335216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello-Fernandez C, Packham G, and Cleveland JL (1993). The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. USA 90, 7804–7808. 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgersdotter A, Sandberg R, and Ernberg I. (2005). Gene expression perturbation in vitro–a growing case for three-dimensional (3D) culture systems. Semin. Cancer Biol 15, 405–412. 10.1016/j.semcancer.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, Lee SH, Lee HR, Myoung J, Ganem D, et al. (2012). Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J. Virol 86, 9708–9720. 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero RA Jr., and Marton LJ (2007). Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat. Rev. Drug Discov 6, 373–390. 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, and Moore PS (1994). Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 266, 1865–1869. 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Choi UY, Lee JJ, Park A, Zhu W, Lee HR, Choi YJ, Yoo JS, Yu C, Feng P, Gao SJ, et al. (2020). Oncogenic human herpesvirus hijacks proline metabolism for tumorigenesis. Proc. Natl. Acad. Sci. USA 117, 8083–8093. 10.1073/pnas.1918607117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffino P. (2001). Regulation of cellular polyamines by antizyme. Nat. Rev. Mol. Cell Biol 2, 188–194. 10.1038/35056508. [DOI] [PubMed] [Google Scholar]
- Coni S, Serrao SM, Yurtsever ZN, Di Magno L, Bordone R, Bertani C, Licursi V, Ianniello Z, Infante P, Moretti M, et al. (2020). Blockade of EIF5A hypusination limits colorectal cancer growth by inhibiting MYC elongation. Cell Death Dis. 11, 1045. 10.1038/s41419-020-03174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leon Vazquez E, and Kaye KM (2011). The internal Kaposi’s sarcoma-associated herpesvirus LANA regions exert a critical role on episome persistence. J. Virol 85, 7622–7633. 10.1128/JVI.00304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker LL, Shankar P, Khan G, Freeman RB, Dezube BJ, Lieberman J, and Thorley-Lawson DA (1996). The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J. Exp. Med 184, 283–288. 10.1084/jem.184.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado T, Sanchez EL, Camarda R, and Lagunoff M. (2012). Global metabolic profiling of infection by an oncogenic virus: KSHV induces and requires lipogenesis for survival of latent infection. PLoS Pathog. 8, e1002866. 10.1371/journal.ppat.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubich T, Dittrich A, Bousset K, Geffers R, Busche G, Koster M, Hauser H, Schulz TF, and Wirth D. (2021). 3D culture conditions support Kaposi’s sarcoma herpesvirus (KSHV) maintenance and viral spread in endothelial cells. J. Mol. Med. (Berl.) 99, 425–438. 10.1007/s00109020-02020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman SH, Ignatenko NA, Powell MB, Blohm-Mangone KA, Holubec H, Guillen-Rodriguez JM, and Gerner EW (1999). APC-dependent changes in expression of genes influencing polyamine metabolism, and consequences for gastrointestinal carcinogenesis, in the Min mouse. Carcinogenesis 20, 1709–1713. 10.1093/carcin/20.9.1709. [DOI] [PubMed] [Google Scholar]
- Fozard JR, Part ML, Prakash NJ, and Grove J. (1980). Inhibition of murine embryonic development by alpha-difluoromethylornithine, an irreversible inhibitor of ornithine decarboxylase. Eur. J. Pharmacol 65, 379–391. 10.1016/0014-2999(80)90342-8. [DOI] [PubMed] [Google Scholar]
- Fujimura K, Wang H, Watson F, and Klemke RL (2018). KRAS oncoprotein expression is regulated by a self-governing eIF5A-PEAK1 feed-forward regulatory loop. Cancer Res. 78, 1444–1456. 10.1158/00085472.CAN-17-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner EW, and Meyskens FL Jr. (2004). Polyamines and cancer: old molecules, new understanding. Nat. Rev. Cancer 4, 781–792. 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Spagnoli GC, Martin I, Ploegert S, Demougin P, Heberer M, and Reschner A. (2005). Three-dimensional culture of melanoma cells profoundly affects gene expression profile: a high density oligonucleotide array study. J. Cell. Physiol 204, 522–531. 10.1002/jcp.20320. [DOI] [PubMed] [Google Scholar]
- Gibson W, van Breemen R, Fields A, LaFemina R, and Irmiere A. (1984). D,L-alpha-difluoromethylornithine inhibits human cytomegalovirus replication. J. Virol 50, 145–154. 10.1128/JVI.50.1.145-154.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco A, Calle A, Morfin F, Thouvenot D, Cayre M, Kindbeiter K, Martin L, Levillain O, and Diaz JJ (2005). S-adenosyl methionine decarboxylase activity is required for the outcome of herpes simplex virus type 1 infection and represents a new potential therapeutic target. FASEB J 19, 1128–1130. 10.1096/fj.04-2108fje. [DOI] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, and Dever TE (2013). eIF5A promotes translation of polyproline motifs. Mol. Cell 51, 35–45. 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke E, Nandigama R, and Ergun S. (2019). Extracellular matrix in the tumor microenvironment and its impact on cancer therapy. Front. Mol. Biosci 6, 160. 10.3389/fmolb.2019.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann D, Conway JR, Vennin C, Magenau A, Hughes WE, Morton JP, and Timpson P. (2014). Three-dimensional cancer models mimic cell-matrix interactions in the tumour microenvironment. Carcinogenesis 35, 1671–1679. 10.1093/carcin/bgu108. [DOI] [PubMed] [Google Scholar]
- Holle AW, Young JL, and Spatz JP (2016). In vitro cancer cell-ECM interactions inform in vivo cancer treatment. Adv. Drug Deliv. Rev 97, 270–279. 10.1016/j.addr.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Huter P, Arenz S, Bock LV, Graf M, Frister JO, Heuer A, Peil L, Starosta AL, Wohlgemuth I, Peske F, et al. (2017). Structural basis for polyproline-mediated ribosome stalling and rescue by the translation elongation factor EF-P. Mol. Cell 68, 515–527.e516. 10.1016/j.molcel.2017.10.014. [DOI] [PubMed] [Google Scholar]
- Ignatenko NA, Babbar N, Mehta D, Casero RA Jr., and Gerner EW (2004). Suppression of polyamine catabolism by activated Ki-ras in human colon cancer cells. Mol. Carcinog 39, 91–102. 10.1002/mc.10166. [DOI] [PubMed] [Google Scholar]
- Ignatenko NA, Gerner EW, and Besselsen DG (2011). Defining the role of polyamines in colon carcinogenesis using mouse models. J. Carcinog 10, 10. 10.4103/1477-3163.79673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby RF, Cole CE, Tutsch K, Newton MA, Kelloff G, Hawk ET, and Lubet RA (2000). Chemopreventive efficacy of combined piroxicam and difluoromethylornithine treatment of Apc mutant Min mouse adenomas, and selective toxicity against Apc mutant embryos. Cancer Res. 60, 1864–1870. [PubMed] [Google Scholar]
- Jensen C, and Teng Y. (2020). Is it time to start transitioning from 2D to 3D cell culture? Front. Mol. Biosci 7, 33. 10.3389/fmolb.2020.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppila S, Stenback F, Risteli J, Jukkola A, and Risteli L. (1998). Aberrant type I and type III collagen gene expression in human breast cancer in vivo. J. Pathol 186, 262–268. . [DOI] [PubMed] [Google Scholar]
- Khan A, Gamble LD, Upton DH, Ung C, Yu DMT, Ehteda A, Pandher R, Mayoh C, Hebert S, Jabado N, et al. (2021). Dual targeting of polyamine synthesis and uptake in diffuse intrinsic pontine gliomas. Nat. Commun 12, 971. 10.1038/s41467-021-20896-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu T, Ballestas ME, Barbera AJ, Kelley-Clarke B, and Kaye KM (2004). KSHV LANA1 binds DNA as an oligomer and residues N-terminal to the oligomerization domain are essential for DNA binding, replication, and episome persistence. Virology 319, 225–236. 10.1016/j.virol.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Lee HR, Choi WC, Lee S, Hwang J, Hwang E, Guchhait K, Haas J, Toth Z, Jeon YH, Oh TK, et al. (2011). Bilateral inhibition of HAUSP deubiquitinase by a viral interferon regulatory factor protein. Nat. Struct. Mol. Biol 18, 1336–1344. 10.1038/nsmb.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy P, and Bartosch B. (2016). Metabolic reprogramming: a hallmark of viral oncogenesis. Oncogene 35, 4155–4164. 10.1038/onc.2015.479. [DOI] [PubMed] [Google Scholar]
- Li J, Meng Y, Wu X, and Sun Y. (2020). Polyamines and related signaling pathways in cancer. Cancer Cell Int. 20, 539. 10.1186/s12935020-01545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Martin HJ, Liao G, and Hayward SD (2007). The Kaposi’s sarcoma-associated herpesvirus LANA protein stabilizes and activates c-Myc. J. Virol 81, 10451–10459. 10.1128/JVI.00804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins JF, Gesteland RF, and Hayashi S. (1995). Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell 80, 51–60. 10.1016/0092-8674(95)90450-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov S, Mailliot J, Rigger L, Neuner S, Shin BS, Yusupova G, Dever TE, Micura R, and Yusupov M. (2016). Molecular insights into protein synthesis with proline residues. EMBO Rep. 17, 1776–1784. 10.15252/embr.201642943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesri EA, Feitelson MA, and Munger K. (2014). Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe 15, 266–282. 10.1016/j.chom.2014.02.011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounce BC, Olsen ME, Vignuzzi M, and Connor JH (2017). Polyamines and their role in virus infection. Microbiol. Mol. Biol. Rev 81. 10.1128/MMBR.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounce BC, Poirier EZ, Passoni G, Simon-Loriere E, Cesaro T, Prot M, Stapleford KA, Moratorio G, Sakuntabhai A, Levraud JP, and Vignuzzi M. (2016). Interferon-induced spermidine-spermine acetyltransferase and polyamine depletion restrict Zika and Chikungunya viruses. Cell Host Microbe 20, 167–177. 10.1016/j.chom.2016.06.011. [DOI] [PubMed] [Google Scholar]
- Myoung J, and Ganem D. (2011). Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods 174, 12–21. 10.1016/j.jviromet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S, and Cleveland JL (2016). Targeting the polyamine-hypusine circuit for the prevention and treatment of cancer. Amino Acids 48, 2353–2362. 10.1007/s00726-016-2275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen ME, Cressey TN, Muhlberger E, and Connor JH (2018). Differential mechanisms for the involvement of polyamines and hypusinated eIF5A in Ebola virus gene expression. J. Virol 92. 10.1128/JVI.0126018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen ME, Filone CM, Rozelle D, Mire CE, Agans KN, Hensley L, and Connor JH (2016). Polyamines and hypusination are required for ebolavirus gene expression and replication. mBio 7. 10.1128/mBio.00882-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH (2006). The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A). J. Biochem 139, 161–169. 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Cooper HL, and Folk JE (1981). Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc. Natl. Acad. Sci. USA 78, 2869–2873. 10.1073/pnas.78.5.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parravicini C, Chandran B, Corbellino M, Berti E, Paulli M, Moore PS, and Chang Y. (2000). Differential viral protein expression in Kaposi’s sarcoma-associated herpesvirus-infected diseases: kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. Am. J. Pathol 156, 743–749. 10.1016/S0002-9440(10)64940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE (2006). Regulation of ornithine decarboxylase. J. Biol. Chem 281, 14529–14532. 10.1074/jbc.R500031200. [DOI] [PubMed] [Google Scholar]
- Pegg AE (2008). Spermidine/spermine-N(1)-acetyltransferase: a key metabolic regulator. Am. J. Physiol. Endocrinol. Metab 294, E995–E1010. 10.1152/ajpendo.90217.2008 . [DOI] [PubMed] [Google Scholar]
- Pegg AE (2009). Mammalian polyamine metabolism and function. IUBMB Life 61, 880–894. 10.1002/iub.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, and Alepuz P. (2017). eIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic Acids Res. 45, 7326–7338. 10.1093/nar/gkx479 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puleston DJ, Baixauli F, Sanin DE, Edwards-Hicks J, Villa M, Kabat AM, Kaminski MM, Stanckzak M, Weiss HJ, Grzes KM, et al. (2021). Polyamine metabolism is a central determinant of helper T cell lineage fidelity. Cell 184, 4186–4202.e4120. 10.1016/j.cell.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Motz M, Jung K, Lassak J, and Frishman D. (2018). Evolutionary analysis of polyproline motifs in Escherichia coli reveals their regulatory role in translation. PLoS Comput. Biol 14, e1005987. 10.1371/journal.pcbi.1005987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom E, and Kahana C. (1994). Polyamines regulate the expression of ornithine decarboxylase antizyme in vitro by inducing ribosomal frame-shifting. Proc. Natl. Acad. Sci. USA 91, 3959–3963. 10.1073/pnas.91.9.3959 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, and Dever TE (2009). Hypusine-containing protein eIF5A promotes translation elongation. Nature 459, 118–121. 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Becker T, Heuer A, Braunger K, Shanmuganathan V, Pech M, Berninghausen O, Wilson DN, and Beckmann R. (2016). Structure of the hypusinylated eukaryotic translation factor eIF-5A bound to the ribosome. Nucleic Acids Res. 44, 1944–1951. 10.1093/nar/gkv1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder S, Hofer SJ, Zimmermann A, Pechlaner R, Dammbrueck C, Pendl T, Marcello GM, Pogatschnigg V, Bergmann M, Muller M, et al. (2021). Dietary spermidine improves cognitive function. Cell Rep. 35, 108985. 10.1016/j.celrep.2021.108985. [DOI] [PubMed] [Google Scholar]
- Schuller AP, Wu CC, Dever TE, Buskirk AR, and Green R. (2017). eIF5A functions globally in translation elongation and termination. Mol. Cell 66, 194–205.e195. 10.1016/j.molcel.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Gan YJ, Davis TO, and Scott RS (2013). Downregulation of the polyamine regulator spermidine/spermine N(1)-acetyltransferase by Epstein-Barr virus in a Burkitt’s lymphoma cell line. Virus Res. 177, 11–21. 10.1016/j.virusres.2013.07.004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood D, Tang-Schomer M, Pouli D, Mizzoni C, Raia N, Tai A, Arkun K, Wu J, Black LD 3rd, Scheffler B, et al. (2019). 3D extracellular matrix microenvironment in bioengineered tissue models of primary pediatric and adult brain tumors. Nat. Commun 10, 4529. 10.1038/s41467019-12420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan V, Komatsu T, Ballestas ME, and Kaye KM (2004). Definition of sequence requirements for latency-associated nuclear antigen 1 binding to Kaposi’s sarcoma-associated herpesvirus DNA. J. Virol 78, 14033–14038. 10.1128/JVI.78.24.14033-14038.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R, Faussat AM, Perrot JY, Dubrulle S, Ramond S, Delmer A, Aj-chenbaum-Cymbalista F, and Marie JP (2004). A new acute lymphoblastic leukaemia cell line BEL-1 with t(4; 11) (q21; q23) chromosomal translocation and a unique aberrant p27 transcript. Br. J. Haematol 126, 754–755. 10.1111/j.1365-2141.2004.05122.x . [DOI] [PubMed] [Google Scholar]
- Tyms AS, and Williamson JD (1982). Inhibitors of polyamine biosynthesis block human cytomegalovirus replication. Nature 297, 690–691. 10.1038/297690a0. [DOI] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, and Jung K. (2013). Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339, 82–85. 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- Wu W, Vieira J, Fiore N, Banerjee P, Sieburg M, Rochford R, Harrington W Jr., and Feuer G. (2006). KSHV/HHV-8 infection of human hematopoietic progenitor (CD34+) cells: persistence of infection during hematopoiesis in vitro and in vivo. Blood 108, 141–151. 10.1182/blood2005-04-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye FC, Zhou FC, Yoo SM, Xie JP, Browning PJ, and Gao SJ (2004). Disruption of Kaposi’s sarcoma-associated herpesvirus latent nuclear antigen leads to abortive episome persistence. J. Virol 78, 11121–11129. 10.1128/JVI.78.20.11121-11129.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappia V, Carteni-Farina M, and Della Pietra G. (1972). S-adenosylmethionine decarboxylase from human prostate. Activation by putrescine. Biochem. J 129, 703–709. 10.1042/bj1290703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw metabolomics data reported in this study are available in the MetaboLights database (https://www.ebi.ac.uk/metabolights/index) under accession numbers MTBLS5218.
The metabolome processed data reported in this study are supplied as Table S1. All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.